Benchmarking Large Language Models for Predictive Modeling in Biomedical Research With a Focus on Reproductive Health
Benchmarking Large Language Models for Predictive Modeling in Biomedical Research With a Focus on Reproductive Health
Sarwal, R.; Tarca, V.; Kalavros, N.; Bhatti, G.; Bhattacharya, S.; Butte, A. J.; Romero, R. J.; Stolovitzky, G.; Oskotsky, T. T.; Tarca, A. L.; Sirota, M.
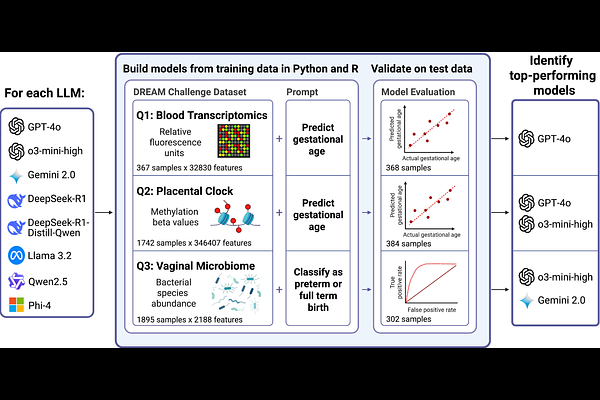
AbstractGenerative AI, particularly large language models (LLMs), is increasingly being used in computational biology to support code generation for data analysis. In this study, we evaluated the ability of LLMs to generate functional R and Python code for predictive modeling tasks, leveraging standardized molecular datasets from several recent DREAM (Dialogue for Reverse Engineering Assessments and Methods) Challenges focused on reproductive health. We assessed LLM performance across four predictive tasks derived from three DREAM challenges: gestational age regression from gene expression, gestational age regression from DNA methylation profiles, and classification of preterm birth and early preterm birth from microbiome data. LLMs were prompted with task descriptions, data locations, and target outcomes. LLM-generated code was then run to fit and apply prediction models and generate graphics, and they were ranked based on their success in completing the tasks and achieving strong test set performance. Among the eight LLMs tested, o3-mini-high, 4o, DeepseekR1 and Gemini 2.0 completed at least one task without error. Overall, R code generation was more successful (14/16 tasks) than Python (7/16), attributed to the utility of Bioconductor packages for querying Gene Expression Omnibus data. OpenAI\'s o3-mini-high outperformed others, completing 7/8 tasks. Test set performance of the top LLM matched or exceeded top-performing teams from the original DREAM challenges. These findings underscore the potential of LLMs to enhance exploratory analysis and democratize access to predictive modeling in omics by automating key components of analysis pipelines, and highlight the potential to increase research output when conducting analyses of standardized datasets from public repositories.