RF-SIRF defining reversed DNA replication forks with single-cell and spatio-temporal resolution reveals a replication stress specific epigenetic code
RF-SIRF defining reversed DNA replication forks with single-cell and spatio-temporal resolution reveals a replication stress specific epigenetic code
Roy, S.; Firmreite, M.; Citron, F.; Draetta, G.; Schlacher, K.
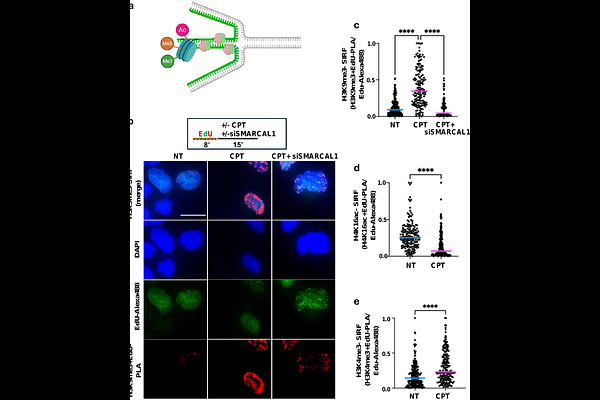
AbstractDNA replication stress responses are key genomic stability guardians critical during development, aging, hematopoiesis, disease suppression and cancer therapy response. Reversed forks (RF) form at stalled DNA replication forks as a distinct four-way DNA structure to protect against formation and exposure to toxic DNA lesions. So far, prevailing methods to measure RFs involve specialized electron microscopy precluding studies within their cellular context. Here we describe an in-situ method to quantitatively measure RFs by harnessing intrinsic bio-physical properties of this distinct DNA structure (RF-SIRF). RF-SIRF reveals that RFs accumulate at the nuclear periphery and form predominantly during early-mid S-phase of the cell cycle. Importantly, RFs are chromatinized and utilize an epigenetic replication stress code distinct from transcription that explains how DNA stress response proteins are recruited to RFs. Collectively, RF-SIRF enables robust, quantitative, temporal and spatial analyses of RFs and associated proteomics, empowering advanced cellular investigations of DNA replication stress responses.