Human O-linked Glycosylation Site Prediction Using Pretrained Protein Language Model
Human O-linked Glycosylation Site Prediction Using Pretrained Protein Language Model
Pakhrin, S. C.; Chauhan, N.; Khan, S.; Upadhyaya, J.; Keller, C.; Neuman, L. N.; Beck, M. R.; Blanco, E.
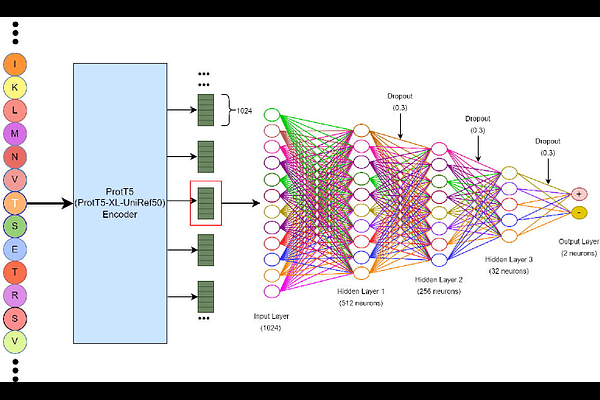
AbstractO-linked glycosylation of proteins is an essential post-translational modification process in Homo sapiens, where the attachment of a sugar moiety occurs at the oxygen atom of serine and/or threonine residues. This modification plays a pivotal role in various biological and cellular functions. While threonine or serine residues in a protein sequence are potential sites for O-linked glycosylation, not all threonine or serine residues are O-linked glycosylated. Furthermore, the modification is reversible. Hence, it is of vital importance to characterize if and when O-linked glycosylation occurs. We propose a multi-layer perceptron-based approach termed OglyPred-PLM which leverages the contextualized embeddings produced from the ProtT5-XL-UniRef50 protein language model that significantly improves the prediction performance of human O-linked glycosylation sites. OglyPred-PLM surpassed the performance of other indispensable O-linked glycosylation predictors on the independent benchmark dataset. This demonstrates that OglyPred-PLM is a powerful and unique computational tool to predict O-linked glycosylation sites in proteins and thus will accelerate the discovery of unknown O-linked glycosylation sites in proteins.