Greater Expression of DNA Repair Pathways in Sharks vs. Rays/Skates Based on Transcriptomic Analyses
Greater Expression of DNA Repair Pathways in Sharks vs. Rays/Skates Based on Transcriptomic Analyses
Simmons, C. R.; Grant, S. L.; Llorente Ruiz, L.; Kerstetter, D. W.; Pimpley, M.; Latimer, J. J.
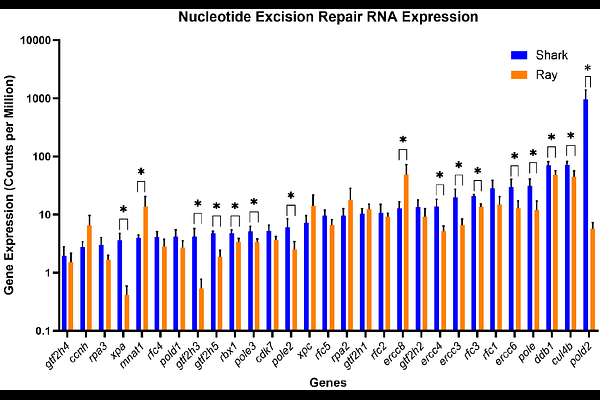
AbstractElasmobranchs are an understudied taxon of cartilaginous fishes. DNA repair studies have been done in very few elasmobranchs. Because DNA repair maintains the integrity of the genetic code, it is important for the survival of elasmobranchs in increasingly polluted oceans. Oil spills, for example, have been shown to cause DNA adducts in ocean creatures. We hypothesized that four elasmobranch species would show differential DNA repair expression. Dermal cells were harvested from nurse sharks (Ginglymostoma cirratum), spiny dogfish (also considered to be sharks) (Squalus acanthias), yellow stingrays (Urobatis jamaicensis), and little skates (Leucoraja erinacea) and RNA was isolated. RNA sequencing was performed using the holocephalan Australian ghostshark (Callorhincus milii) reference genome. RNA samples from the sharks manifested significantly higher expression than those of rays/skates in 4/5 major DNA repair pathways (Base Excision, Nucleotide Excision, Mismatch Repair, and Homologous Recombination). Each of the four pathways of DNA repair manifested differential expression of pathway-specific genes (xpa, gtf2h3, mpg, rad51c,d, mlh1, ercc3, ercc6, xrcc3, parp1, atr, brip1, top3A, blm, ddb1, fancd2, etc.). One gene, polD2, was consistently elevated in the sharks vs. rays/skates in all four pathways. Although we studied RNA from skin, polD2, a subunit of polD, has been proposed to contribute to the spreading and amplification of hypermutations in sharks by generating higher diversity of the T cell receptor repertoire. In summary, with increased expression of four major DNA repair pathways, sharks may be more successful than rays/skates in surviving the genotoxic effects of increasingly polluted oceans.