Expanding the Orchidaceae Virome Through Analysis of Public-Domain Sequencing Data
Expanding the Orchidaceae Virome Through Analysis of Public-Domain Sequencing Data
Perez, A.; Sanchez-Yali, J.; Higuita, M.; Ortiz-Reyes, A.; Gutierrez, P. A.
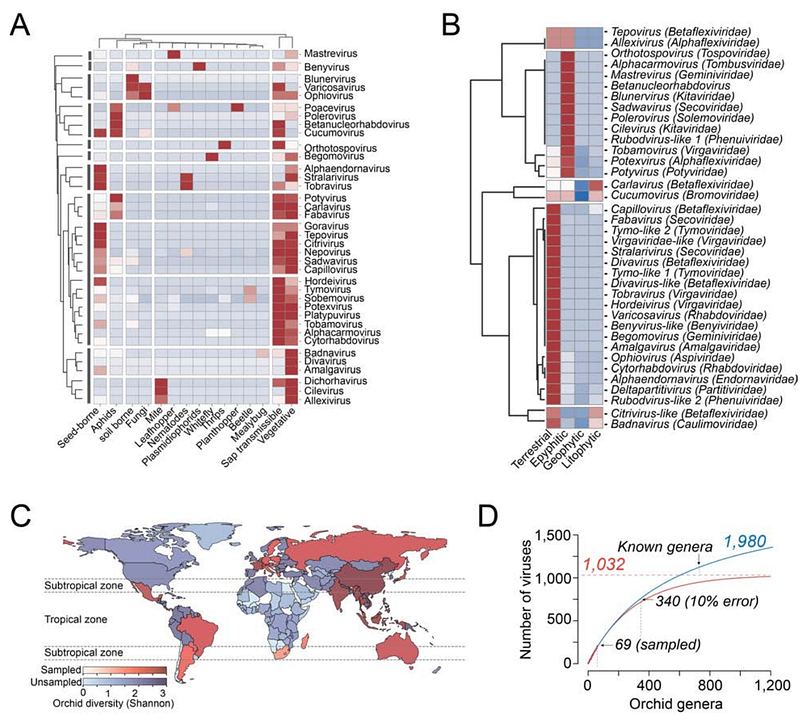
AbstractOrchidaceae is one of the largest plant families, renowned for its ornamental appeal, uses in food, cosmetics, and medicine, and its vital importance in ecosystem health and biodiversity. However, despite their diversity and economic significance, knowledge about the viruses associated with orchid species remains limited. Fortunately, the widespread adoption of high-throughput sequencing enables the collection of indirect information about the viruses associated with various plant hosts. In this work, we conducted an integrated data analysis of viruses associated with Orchidaceae by reexamining 2,789 public RNA-seq datasets encompassing 41 genera, 18 subtribes, and five subfamilies. We identified sequences associated with 163 viruses, 13 representing new associations with Orchidaceae. Additionally, 117 sequences likely represent putative novel species, including six tentative new genera within Benyviridae, Betaflexiviridae, Phenuiviridae, and Tymoviridae. Analysis of the updated Orchidaceae virome revealed a strong correlation between the pairs Alphaflexiviridae/Virgaviridae, Rhabdoviridae/Partitiviridae, and Partitiviridae/Secoviridae and a negative correlation between Alphaflexiviridae/Partitiviridae; Virgaviridae/Partitiviridae; Rhabdoviridae/Alphaflexiviridae; Betaflexiviridae/Alphaflexiviridae; and Secoviridae/Alphaflexiviridae in addition to correlations between virus genera and orchid lifestyles. A network analysis of the host range of orchid viruses was also performed in addition to a preliminary estimate of the size of the Orchidaceae virome. IMPORTANCEDespite their diversity, little is known about viruses associated with orchids. Plant viruses significantly threaten agriculture and may harm orchid conservation. Without detailed characterization of the orchid virome and suitable diagnostic tools, viruses could unintentionally enter ecosystems and cultivation fields from contaminated sources. Sequence information is crucial for developing detection tests for quick virus identification, supporting disease management strategies, and producing virus-free plants. In addition, understanding ecological factors contributing to virus transmission--like primary infection sources, vectors, and alternative hosts--is essential for selecting control measures to prevent viral diseases and manage outbreaks. Data mining provides a cost-effective method to bridge knowledge gaps, highlight trends that are useful for plant virus surveillance, and inform epidemiological decisions. The analysis presented here offers a global perspective on the Orchidaceae virome that could guide future research and virus vigilance programs and enhance our understanding of plant virus diversity.