ClaPNAC: a Classifier of Protein - Nucleic Acid Contacts
ClaPNAC: a Classifier of Protein - Nucleic Acid Contacts
Nikolaev, G.; Baulin, E. F.; Bujnicki, J. M.
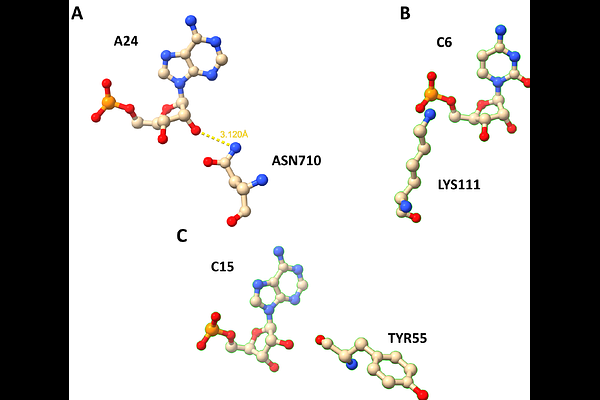
AbstractNucleic acid-protein interactions are fundamental to many cellular processes, including genome stability and replication, and the regulation of gene expression. They rely on the formation of specific contacts between amino acid and nucleotide residues. Accurate detection and classification of these inter-residue contacts in both experimentally determined structures and computationally predicted models of nucleic acid-protein complexes are crucial for achieving a comprehensive, molecular-level understanding of nucleic acid-protein recognition mechanisms. Here, we present ClaPNAC, a classifier that annotates 3D structures of nucleic acid-protein complexes with pairwise contacts. It distinguishes stacking interactions (involving the faces of nucleobases), pseudo pairs (involving the Watson-Crick, Hoogsteen, or sugar edge of the base), and phosphate and ribose interactions on the nucleoside/nucleotide side, as well as side chain and backbone interactions on the amino acid side. ClaPNAC is based on a geometric approach, extending our previous method ClaRNA and uses a database of pre-classified doublets extracted from experimentally solved RNA-protein complex structures. Unlike other tools that focus on specific chemical interactions such as hydrogen bonds or van der Waals contacts, ClaPNAC relies on the geometry of the interacting residues. It is applicable to both RNA- and DNA-protein complexes, and it can also operate on isolated nucleosides/nucleotides and amino acids. It can be extended to support additional interaction types and higher-order spatial relationships. ClaPNAC constitutes a notable advancement in the characterization of amino acid-nucleotide interactions by providing a comprehensive classification framework that deepens our insight into these fundamental biological processes. Its applicability to 3D structure prediction and validation highlights its value as a resource for researchers in structural biology.