Eco-evolutionary Guided Pathomic Analysis to Predict DCIS Upstaging
Eco-evolutionary Guided Pathomic Analysis to Predict DCIS Upstaging
Xiao, Y.; Elmasry, M.; Bai, J.; Chen, A.; Chen, Y.; Jackson, B.; Johnson, J. O.; Gillies, R. J.; Prasanna, P.; Chen, C.; Damaghi, M.
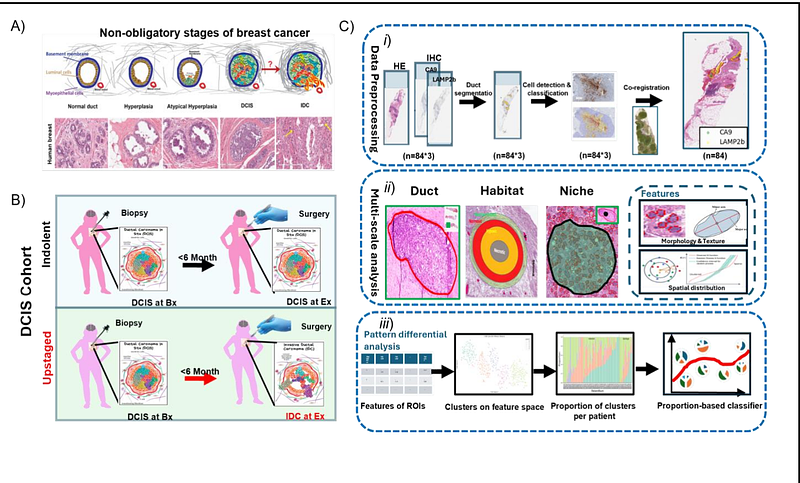
AbstractCancers evolve in a dynamic ecosystem. Thus, characterizing cancer\'s ecological dynamics is crucial to understanding cancer evolution and can lead to discovering novel biomarkers to predict disease progression. Ductal carcinoma in situ (DCIS) is an early-stage breast cancer characterized by abnormal epithelial cell growth confined within the milk ducts. Although there has been extensive research on genetic and epigenetic causes of breast carcinogenesis, none of these studies have successfully identified a biomarker for the progression and/or upstaging of DCIS. In this study, we show that ecological habitat analysis of hypoxia and acidosis biomarkers can significantly improve prediction of DCIS upstaging. First, we developed a novel eco-evolutionary designed approach to define habitats in the tumor intra-ductal microenvironment based on oxygen diffusion distance in our DCIS cohort of 84 patients. Then, we identify cancer cells with metabolic phenotypes attributed to their habitat conditions, such as the expression of CA9 indicating hypoxia responding phenotype, and LAMP2b indicating a hypoxia-induced acid adaptation. Traditionally these markers have shown limited predictive capabilities for DCIS upstaging, if any. However, when analyzed from an ecological perspective, their power to differentiate between indolent and upstaged DCIS increased significantly. Second, using eco-evolutionary guided computational and digital pathology techniques, we discovered distinct spatial patterns of these biomarkers and used the distribution of such patterns to predict patient upstaging. The patterns were characterized by both cellular features and spatial features. With a 5-fold validation on the biopsy cohort, we trained a random forest classifier to achieve the area under curve(AUC) of 0.74. Our results affirm the importance of using eco-evolutionary-designed approaches in biomarkers discovery studies in the era of digital pathology by demonstrating the role of eco-evolution dynamics in predicting cancer progression.


