Merging conformational landscapes in a single consensus space with FlexConsensus algorithm
Merging conformational landscapes in a single consensus space with FlexConsensus algorithm
Herreros, D.; Perez Mata, C.; Sorzano, C. O. S.; Carazo, J. M.
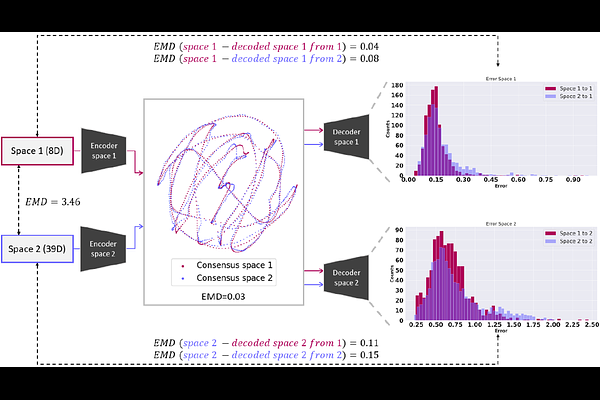
AbstractThe analysis of structural heterogeneity in CryoEM is experimenting a significant breakthrough toward estimating more accurate, richer, and interpretable conformational landscapes directly derived from experimental data. The increasing number of new methods designed to tackle the heterogeneity challenge reflects these new paradigms, allowing users to understand protein dynamics better. However, a question remains on how the estimation of different heterogeneity algorithms compare, which is essential to properly determine the reliability, stability, and correctness of the conformational landscapes and the structural transitions extracted from the CryoEM images. The intrinsic differences arising from the approximation and implementation of every heterogeneity algorithm make the definition of a consensus complex, a problem that remains unsolved. To overcome the challenges of comparing heterogeneity algorithms, we introduce our new FlexConsenus algorithm in this work. FlexConsensus relies on a multi-autoencoder architecture to learn the commonalities and differences of several conformational landscapes, allowing one to place them in a shared consensus space with enhanced reliability. In addition, FlexConsensus enables the derivation of error metrics from the consensus space to measure the reproducibility of the heterogeneity estimations. Thanks to the previous consensus metrics, it is possible to clean the original particle datasets further based on the differences in the estimation of their structural variability among different algorithms and/or different runs, allowing users to focus their analysis only on those particles with a stable estimation of their structural variability.