Ultraconserved elements coupled with machine learning approaches resolve the systematics in model nematode species
Ultraconserved elements coupled with machine learning approaches resolve the systematics in model nematode species
Villegas, L. I.; Jimenez, L.; van der Sprong, J.; Holovachov, O.; Waldvogel, A.-M.; Schiffer, P. H.
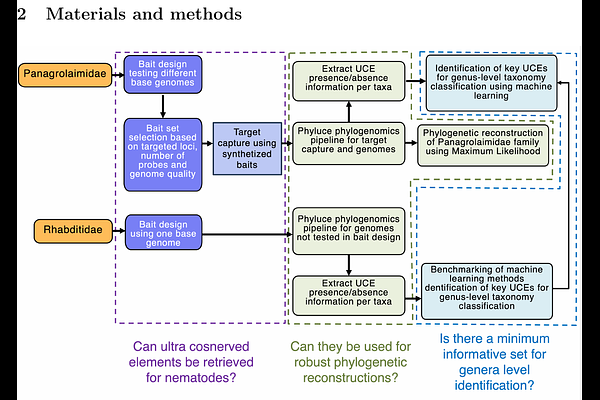
AbstractNematodes are among the most diverse animal groups, inhabiting nearly all terrestrial and aquatic ecosystems. More than a million species of nematodes are expected to occur on Earth. However, only around 28,000 have been described to date. Nematode phylogenetics remains challenging due to their small size, morphological simplicity, and cryptic diversity. Traditional morphological and molecular approaches, such as single-locus markers (e.g., 18S rRNA, COI), often lack resolution, particularly at shallow evolutionary scales. Moreover, morphology-based classifications are flawed, and phylogenies inferred from single-locus data often fail to resolve deep branching relationships within the nematode phylogenetic tree. These limitations highlight the need for more comprehensive genomic approaches to achieve higher-resolution evolutionary inferences. Here, we design and test the first ultraconserved elements (UCEs) probe set for two nematode families: Panagrolaimidae and Rhabditidae. This approach captures thousands of loci without requiring whole-genome or transcriptome sequencing. Our probe sets targeted 1612 and 100397 UCE loci for Panagrolaimidae and Rhabditidae, respectively. In vivo testing for Panagrolaimidae captured up to 1457 loci, enabling a robust phylogenetic reconstruction. Genera classifications within Panagrolaimidae were congruent with prior phylogenies, except for one strain, which we here redescribe as Neocephalobus halophilus BSS8, based on morphological and molecular evidence. We applied machine learning classifiers to determine the minimum loci required for genus-level classification. For the Rhabditidae family, benchmarking of machine learning models revealed that XGBoost provided the highest accuracy for genus-level classification, with 46 loci as the most informative. For Panagrolaimidae, despite the availability of extensive laboratory isolates, genomic resources are limited. We identified 63 loci as the most informative for the classification of this family. In summary, our UCE probe sets provide a scalable and cost-effective tool for enhancing taxonomic resolution and evolutionary inference in nematodes. This approach has the potential to improve biodiversity assessments and deepen our understanding of this ecologically important group, even with shallow sequencing approaches on-site.