Ammonia-mediated Conformational Dynamics in SCAP for SREBP Activation
Ammonia-mediated Conformational Dynamics in SCAP for SREBP Activation
Chan, C.; Li, Z.; Yang, Y.; Guo, D.; Cheng, X.
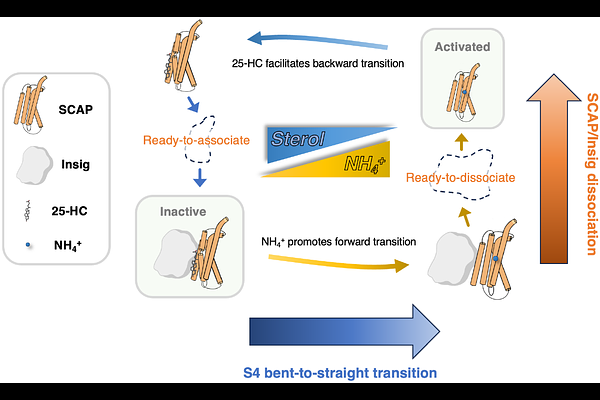
AbstractCholesterol homeostasis is regulated by the sterol regulatory element-binding protein (SREBP) pathway, with the membrane-embedded SREBP-cleavage-activating protein (SCAP) and insulin-inducible gene protein (Insig)-1/2 acting as sterol sensors. Our previous studies unveiled a critical role of ammonia in activating the SREBP pathway. Here, we performed extensive molecular dynamics (MD) simulations to elucidate the structural mechanisms through which ammonia binding triggers SCAP/Insig dissociation and subsequently activate SREBP. The SCAP S4 helix exists in two distinct conformations: a partially unfolded S4 and an intact S4; the transition between these states may constitute a crucial aspect of SCAP activation. We first studied the binding of ammonia to SCAP in different S4 conformational states. We then explored how the binding of ammonia or 25-hydroxycholesterol (25-HC) affects the conformational transition of S4 using targeted MD simulations. Finally, we performed comparative analysis of the 25-HC-bound inactive SCAP and the ammonia-bound active SCAP. Collectively, our findings highlight the importance of S4 helix conformation in activating the SCAP protein. Our simulations suggest that ammonia and 25-HC act as an agonist and antagonist of SCAP activation, respectively. Specifically, ammonia binding facilitates the transition of S4 from the unfolded to the straight conformation, triggering subsequent conformational changes in the transmembrane domain of SCAP. These changes eventually promote the SCAP/Insig dissociation and alter the position of the MELADL motif at the membrane surface.