VirusImmu: a novel ensemble machine learning approach for viral immunogenicity prediction
VirusImmu: a novel ensemble machine learning approach for viral immunogenicity prediction
Li, J.; Zhao, Z.; Tai, C.; Sun, T.; Tan, L.; Li, X.; He, W.; Li, H.; Zhang, J.
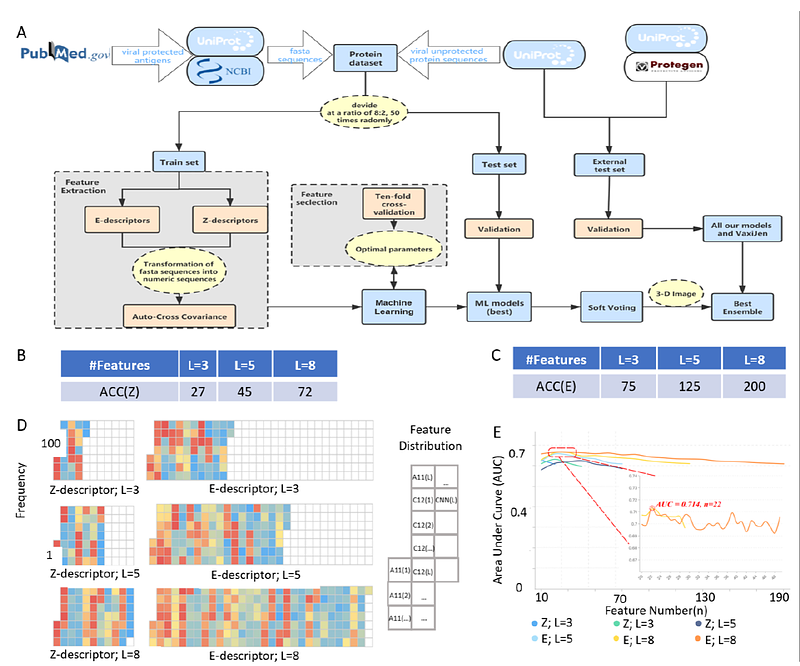
AbstractBackground: The viruses threats provoke concerns regarding their sustained epidemic transmission, making the development of vaccines particularly important. In the prolonged and costly process of vaccine development, the most important initial step is to identify protective immunogens. Machine learning (ML) approaches are productive in analyzing big data such as microbial proteomes, and can remarkably reduce the cost of experimental work in developing novel vaccine candidates. Results: We intensively evaluated the immunogenicity prediction power of eight commonly-used ML methods by random sampling cross validation on a large dataset consisting of known viral immunogens and non-immunogens we manually curated from the public domain. XGBoost, kNN and RF showed the strongest predictive power. We then proposed a novel soft-voting based ensemble approach (VirusImmu), which demonstrated a powerful and stable capability for viral immunogenicity prediction across the test set and external test set irrespective of protein sequence length. VirusImmu was successfully applied to facilitate identifying linear B cell epitopes against African Swine Fever Virus as confirmed by indirect ELISA in vitro. Conclusions: VirusImmu exhibited tremendous potentials in predicting immunogenicity of viral protein segments. It is freely accessible at https://github.com/zhangjbig/VirusImmu.