Design of overlapping genes using deep generative models of protein sequences
Design of overlapping genes using deep generative models of protein sequences
Byeon, G. W.; Exposit, M.; Baker, D.; Seelig, G.
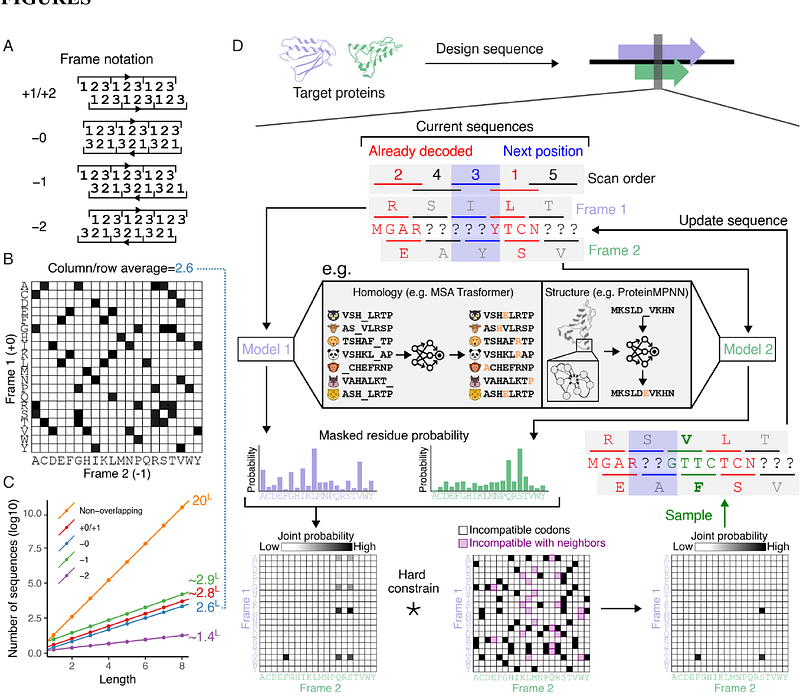
AbstractIn nature, viruses frequently evolve overlapping genes (OLG) in alternate reading frames of the same nucleotide sequence despite the drastically reduced protein sequence space resulting from the sharing of codon nucleotides. Their existence leads one to wonder whether amino acid sequences are sufficiently degenerate with respect to protein folding to broadly allow arbitrary pairs of functional proteins to be overlapped. Here, we investigate this question by engineering synthetic OLGs using state-of-the-art generative models. To evaluate the approach, we first design overlapped sequences targeting two different protein families. We then encode distinct highly ordered de novo protein structures and observe surprisingly high in silico and experimental success rates. This demonstrates that the overlap constraints under the structure of the standard genetic code do not significantly restrict simultaneous accommodation of well defined 3D folds in alternative reading frames. Our work suggests that OLG sequences may be frequently accessible in nature and could be readily exploited to compress and constrain synthetic genetic circuits.