Differential role of phosphorylation in glucagon family receptor signaling revealed by mass spectrometry
Differential role of phosphorylation in glucagon family receptor signaling revealed by mass spectrometry
Lamb, I. M.; White, A. D.; Willard, F. S.; Chalmers, M. J.; Xiao, J. M.
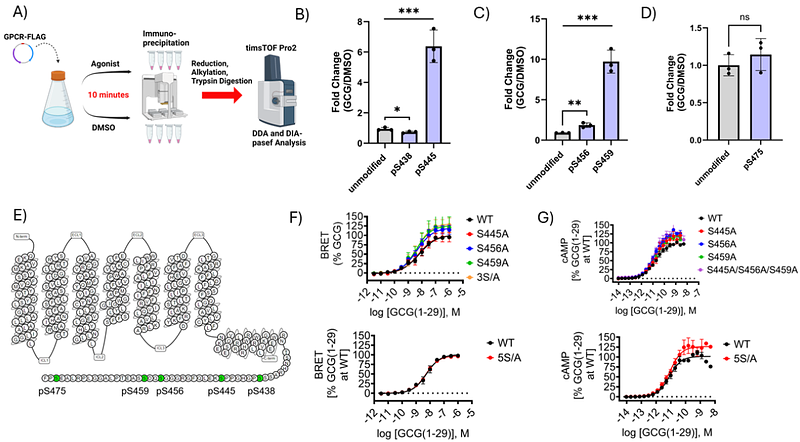
AbstractIn response to extracellular ligands, G protein-coupled receptors (GPCRs) undergo conformational changes that induce coupling to intracellular effectors such as heterotrimeric G proteins that trigger various downstream signaling pathways 1. These events have been shown to be highly regulated by the concerted effects of post-translational modifications (PTMs) that occur in a ligand-dependent manner. Most notably, phosphorylation of residues in the C-terminal cytoplasmic tail of GPCRs has been strongly implicated in promoting receptor interactions with {beta}-arrestins ({beta}arrs) 2, 3 which are cytosolic adaptor proteins that modulate G protein coupling 4, receptor internalization 5, and perhaps also serve as signaling modules in their own right 6. Here, we use proteomic methods to identify C-tail residues that are phosphorylated in the glucagon family of class B1 GPCRs (GLP-1R, GCGR, GIPR) upon agonist addition. Mutagenesis studies reveal unique effects of phosphorylated residues on {beta}arr recruitment and cyclic AMP (cAMP) production that occur in a receptor-dependent manner. We demonstrate that phosphorylation of GLP-1R and GIPR is a critical determinant in driving the formation of GPCR-{beta}arr complexes. However, our results suggest that ligand-induced {beta}arr recruitment to the GCGR proceeds in a phosphorylation-independent manner. These findings highlight the importance of recognizing phosphorylation as a component in the regulation of class B1 GPCR signaling, but also the need to consider how such phenomena may not necessarily yield identical effects on intracellular signaling cascades.