A Computational Framework for Analysis of cfDNA Fragmentation Profiles
A Computational Framework for Analysis of cfDNA Fragmentation Profiles
Poh, Z. W.; Zhu, G.; Wong, P. M.; Odinokov, D.; Carrie, H.; Lau, Y. T.; Gan, A.; Poon, P.; Tan, P.; Jacobsen Skanderup, A.; Tan, I.
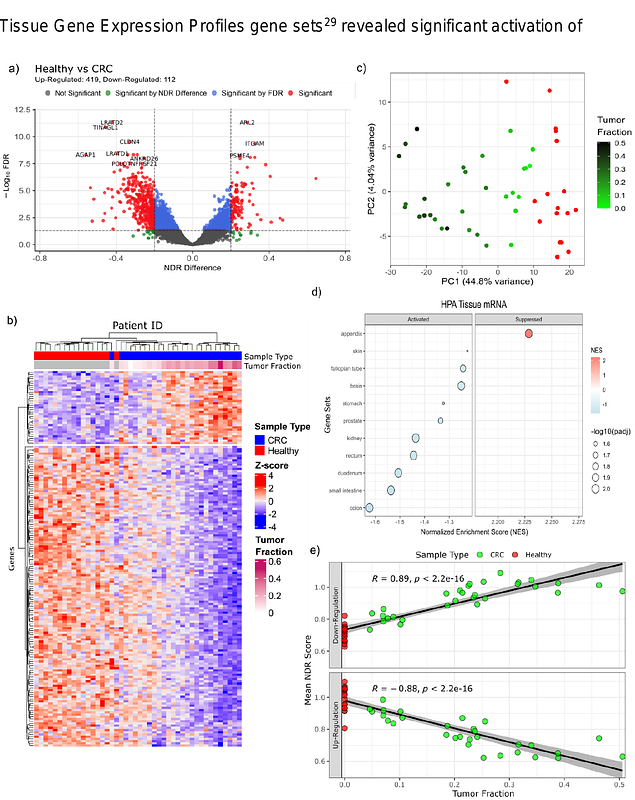
AbstractCirculating cell-free DNA (cfDNA) has emerged as a promising non-invasive medium for studying tumor molecular profiles. Non-random fragmentation patterns in plasma cfDNA, particularly around nucleosome-depleted regions (NDRs) near transcription start sites (TSS), have been shown to reflect epigenetic regulation and gene expression. In this study, coverage profiles of the NDR were utilized to derive an NDR score, which was subsequently used as a proxy for inferring gene expression. To reduce transcript-to-transcript variability and enhance the clarity of these expression-associated signals, we implement a method for GC-bias correction of cfDNA samples. A computational framework (NDRDiff) was then developed to enable comparative analyses of NDR score profiles across different sample groups. The GC-bias correction preserved the overall trend of the NDR signal while improving the separation of gene expression levels, as demonstrated by comparisons of healthy donor cfDNA samples with matched blood RNA-seq data. Validation on a simulated dataset showed that NDRDiff achieved an area under the precision-recall curve (AUPRC) of 0.916, outperforming a standard t-test (AUPRC of 0.777). When applied to a comparison of healthy donor cfDNA and metastatic colorectal cancer (mCRC) cfDNA, NDRDiff identified 531 differential NDR score (DNS) genes that facilitated clear separation between the two groups. These DNS genes were found to correlate with tumor fraction estimates (down-regulated DNS genes: Pearson R = 0.89, p < 0.05; up-regulated DNS genes: Pearson R = -0.88, p < 0.05) and included CLDN4, BIN2, and IRAG2, which exhibit strong associations with colorectal cancer or blood cell expression signatures. Gene set enrichment analysis further revealed enrichment of colon and other gastrointestinal tissue signatures. Collectively, these findings underscore the potential of NDR-based cfDNA analysis as a minimally invasive tool for monitoring tumor-related molecular features in cancer.