On interactive spatial visualisation of pathogenicity predictions
On interactive spatial visualisation of pathogenicity predictions
Xu, J.; Kovacs, A. S.; Portelli, S.; Malik, A. J.; Ascher, D. B.
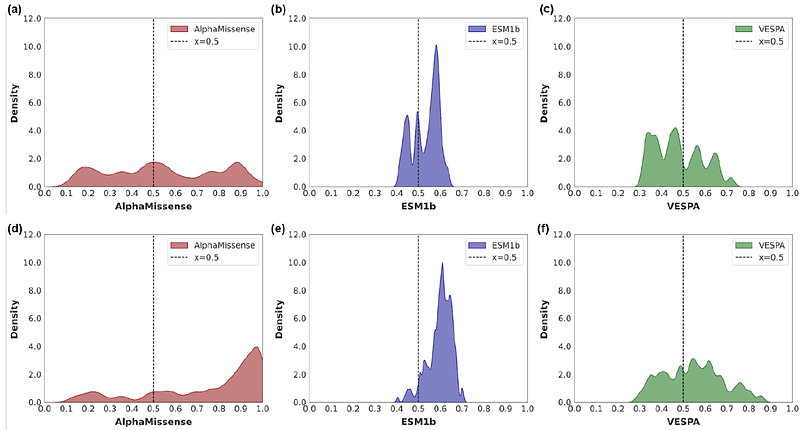
AbstractThe past few years have revolutionised the utility of protein structural information in assessing disease risk. Such efforts have been pioneered by the DeepMind and Meta teams, who initially developed accurate protein prediction tools AlphaFold and ESMFold, respectively. More recently, both have been readapted to create AlphaMissene and ESM1b, through which, the pathogenicity potential of all possible missense mutations within the human proteome has been predicted. Similarly, VESPA was created as an alignment-agnostic predictor, which infers conservation from protein-language model embeddings to assign an effect for each variant. To facilitate their utility, we present a unified dashboard, MissenseViewer, which permits direct predictor comparison, and visualisation within the protein 3-dimensional structural context. Our interactive resource is freely available at: https://biosig.lab.uq.edu.au/missenseviewer/.