Evolutionary consequences of bacterial resistance to a flagellotropic phage
Evolutionary consequences of bacterial resistance to a flagellotropic phage
Antani, J. D.; Theroux, A.; Emonet, T.; Turner, P. E.
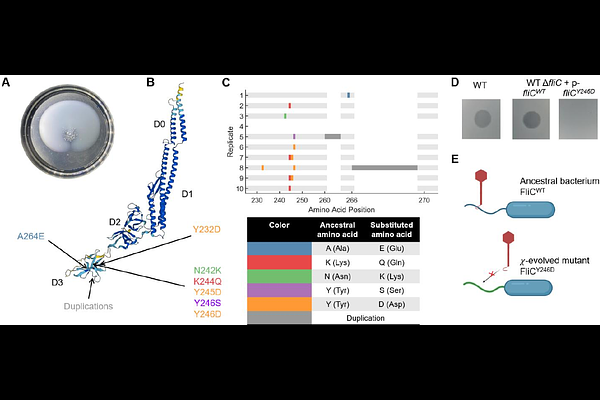
AbstractBacteria often rapidly evolve resistance to bacteriophages (phages) by mutating or suppressing the phage-receptors, the factors that phages first target to initiate infection. Flagellotropic phages infect bacteria by initially binding to the flagellum. Since motility is an important fitness factor that allows bacteria to efficiently explore their environment, losing flagellar function to evade infection by flagellotropic phages represents a crucial trade-off. In this study, we investigated the evolutionary responses of Escherichia coli when exposed to the flagellotropic phage {chi}. Using an experimental evolution approach, E. coli cells were repeatedly subjected to environments rich in phage {chi} but selective for motility. Unlike traditional well-mixed cultures, we employed swim-plate assays to simulate spatial confinement and promote motility. Whole genome sequencing of evolved populations revealed early emergence of non-motile, {chi}-resistant mutants with mutations disrupting motility-related genes. Motile mutants emerged in later passages, possessing mutations in the flagellin gene fliC. Swim-plate assays showed a diverse range of motility among these mutants, with some displaying slower, and others faster, expansion speeds compared to the ancestral strain. Single-cell tracking experiments indicated an increased tumble bias in {chi}-resistant mutants, suggesting an adaptive response involving altered flagellar rotation. Our findings demonstrate that motility can undergo trade-offs and trade-ups with phage resistance, shedding light on the complex evolutionary dynamics between motile bacteria and flagellotropic phages.