Functional metagenomics reveals an alternative, broad-specificity pathway for metabolism of carbohydrates in human gut commensal bacteria
Functional metagenomics reveals an alternative, broad-specificity pathway for metabolism of carbohydrates in human gut commensal bacteria
Nasseri, S. A.; Lazarski, A. C.; Lemmer, I. L.; Zhang, C. Y.; Brencher, E.; Chen, H.-M.; Sim, L.; Betschart, L.; Worral, L. J.; Strynadka, N. C. J.; Withers, S. G.
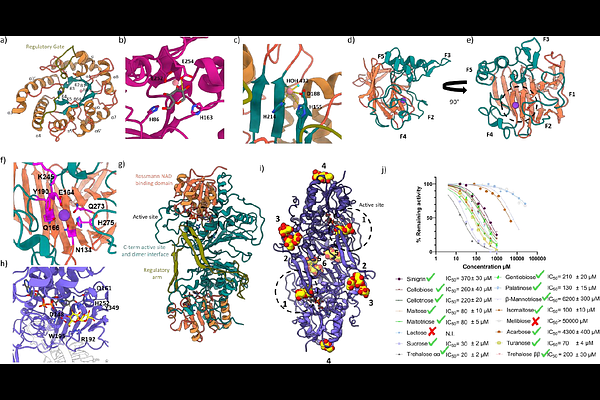
AbstractThe vast majority of the glycosidases characterised so far follow one of the variations of the 'Koshland' mechanisms to hydrolyse glycosidic bonds. Herein we describe a large-scale screen of a human gut microbiome metagenomic library using an assay that selectively identifies non-Koshland glycosidase activities. This screen led to identification of a commonly occurring cluster of enzymes with unprecedentedly broad substrate specificities that is thoroughly characterised, mechanistically and structurally. Not only do these enzymes break glycosidic linkages of both and {beta} stereochemistry and multiple connectivities, but also substrates that are not cleaved by standard glycosidases. These include thioglycosides such as glucosinolates and pseudo-glycosidic bonds of pharmaceuticals such as acarbose. This is achieved via a distinct mechanism of hydrolysis that involves stepwise oxidation, elimination and hydration steps, each catalysed by enzyme modules that are in many cases interchangeable between organisms and substrate classes. These appear to constitute a substantial alternative pathway for glycan degradation.