A Dose-Response Model for Accurate Detection and Quantification of Transcriptome-Wide Gene Knockdown for Oligonucleotide-Based Medicines
A Dose-Response Model for Accurate Detection and Quantification of Transcriptome-Wide Gene Knockdown for Oligonucleotide-Based Medicines
Pekker, D.; Kuntz, S.; Nicholson-Shaw, T.; Yanke, S.; Mukhopadhyay, S.; McArthur, M.
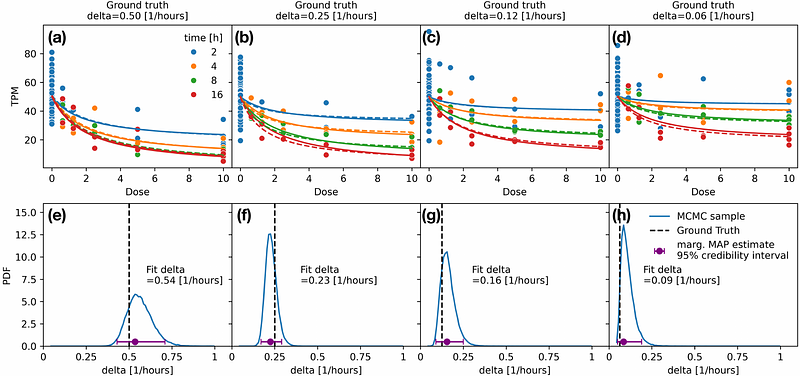
AbstractSynthetic antisense oligonucleotides and siRNAs are a class of Oligonucleotide-Based Medicines (OBMs) that can hybridize with pre-mRNA and mRNA, recruit a mechanism-of-action specific enzymatic complex, and knockdown target gene expression. This class of molecules provides an excellent substrate for designing precision gene-modulatory therapeutics; however, quantifying on- and off-target dose response as measured by next-generation sequencing for this class of therapeutics has remained under-powered and ambiguous. Often in silico predictions of off-targets (ranked by edit tolerance) are used as putative off-target analysis in ASO and siRNA drug design. We construct a simple, effective theory of transcriptional dynamics and enzymatic activity in order to describe the transcriptome-wide response to these oligonucleotides. We establish rigorous quantification methods of off-target analysis in oligonucleotide drug design. We also extend the DESeq work of Negative Binomial noise in gene expression measurements to describe noise, including outliers, in OBM-dose response NGS experiments. We demonstrate the performance of our model on both synthetic and experimental Digital Gene Expression (DGE) data of dose response in ASO-treated cells. We present our analysis package, DoReSeq, as a freely available resource for the community. We hope this will elevate the standards of off-target analysis for such an important class of precision therapeutics.


