Hybracter: Enabling Scalable, Automated, Complete and Accurate Bacterial Genome Assemblies
Hybracter: Enabling Scalable, Automated, Complete and Accurate Bacterial Genome Assemblies
Bouras, G.; Houtak, G.; Wick, R. R.; Mallawaarachchi, V.; Roach, M. J.; Papudeshi, B.; Judd, L. M.; Sheppard, A. E.; Edwards, R. A.; Vreugde, S.
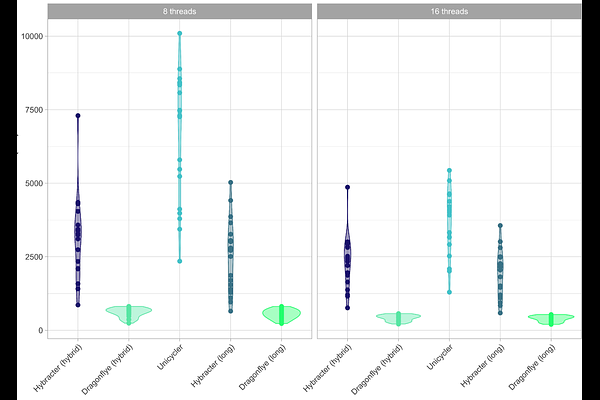
AbstractImprovements in the accuracy and availability of long-read sequencing mean that complete bacterial genomes are now routinely reconstructed using a long-read first assembly approach, usually supplemented with short-read polishing. Complete genomes allow a deeper understanding of bacterial evolution and genomic variation beyond small nucleotide variants (SNVs). They allow for the detection of larger structural variants and the identification of plasmid sequences distinct from the chromosome, which often carry medically significant antimicrobial resistance (AMR) genes. Here, we present Hybracter, a fast and scalable tool implemented in Snakemake with a Snaketool-powered command line interface that allows fast automatic recovery of near-perfect complete bacterial genomes. We compared Hybracter to other contemporary automated command-line hybrid assembly tools using reads from a panel of isolates with matching manually curated genome assemblies as ground truth references. We demonstrate that Hybracter is significantly more accurate and faster than the current popular gold standard automated hybrid assembler Unicycler.