Hybrid Deep Learning with Protein Language Models and Dual-Path Architecture for Predicting IDP Functions
Hybrid Deep Learning with Protein Language Models and Dual-Path Architecture for Predicting IDP Functions
Liang, J.; Peng, Z.
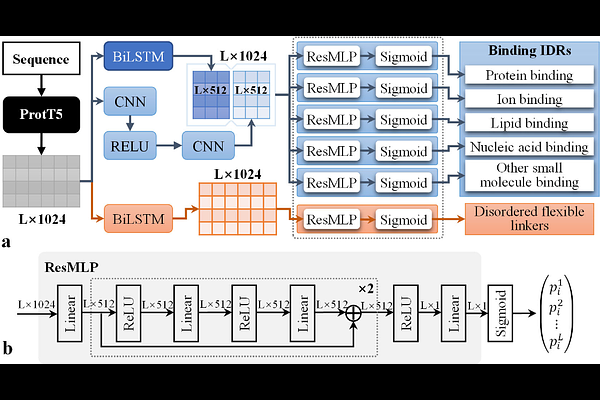
AbstractIntrinsically disordered regions (IDRs) drive essential cellular functions but resist conventional structural-function annotation due to their dynamic conformations. Current computational methods struggle with cross-dataset generalization and functional subtype discrimination. We present IDPFunNet, a hybrid deep learning model combining convolutional neural network, bidirectional LSTM, residual MLP, and the protein language model ProtT5 to predict six IDR functional classes: five binding subtypes and disorded flexible linkers (DFLs). Its dual-path architecture decouples binding IDR prediction and DFL identification using evolutionary embeddings from ProtT5, which outperformed ESM-family models and AlphaFold2 structural features by [≥]1.3% average AUC and [≥]12.7% average APS. Validated across six benchmarks including CAID2/3 blind tests, IDPFunNet achieves superior protein-binding prediction (AUC 0.832-0.866) with significant improvements ([≥]1.5%, p-value<0.05) over existing methods, while matching specialized DFL predictors. Multi-task learning enhances protein/lipid/small molecule-binding performance (3.1-35.1% AUC gains) with BiLSTMs proving optimal DFL identification, despite self-attention\'s potential for nucleic acid-binding (AUC 0.831). This framework overcomes methodological limitations through interpretable, generalizable IDR functional mapping.