Integrative analysis of mRNA stability regulation uncovers a metastasis-suppressive program in breast cancer
Integrative analysis of mRNA stability regulation uncovers a metastasis-suppressive program in breast cancer
Karner, H.; Mittmann, T. C.; Chen, V. W.; Borah, A. A.; Langen, A.; Yousefi, H.; Fish, L.; Zaro, B. W.; Navickas, A.; Goodarzi, H.
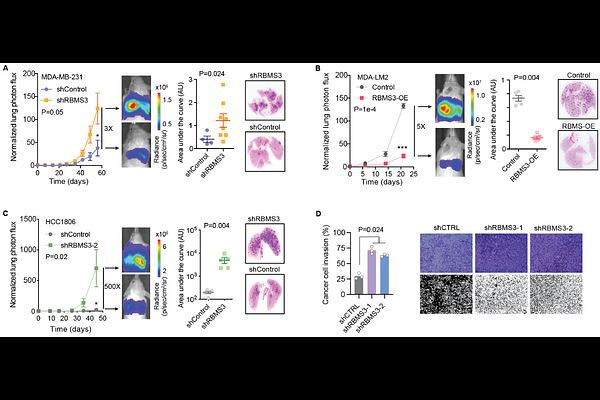
AbstractHeterogeneity in cancer gene expression is typically linked to genetic and epigenetic alterations, yet post-transcriptional regulation likely influences these patterns as well. However, the quantitative contribution of post-transcriptional mechanisms to cancer transcriptome dynamics remains unclear. Here, we systematically measured mRNA dynamics across diverse breast cancer models, revealing that mRNA stability significantly shapes gene expression variability. To decipher the regulatory grammar underlying these dynamics, we developed GreyHound, an interpretable multimodal deep-learning framework integrating RNA sequence features and RNA-binding protein (RBP) expression. GreyHound identified an extensive network of RBPs and their regulons underlying variations in mRNA stability. Among these, we uncovered a metastasis-suppressive regulatory axis centered on the RNA-binding protein RBMS3 and its post-transcriptional control of the redox regulator TXNIP. Functional and molecular analyses revealed that RBMS3 depletion resulted in targeted transcript destabilization, which was associated with poor clinical outcomes and enhanced metastatic potential in xenograft models. Using in vivo epistasis studies, we confirmed that RBMS3 regulation of TXNIP mRNA stability drives this metastasis-suppressive program. These findings position the RBMS3-TXNIP regulatory axis as a key post-transcriptional mechanism in breast cancer and illustrate how interpretable models of RNA dynamics can uncover hidden regulatory programs in disease.