Creation and validation of LIMON - Longitudinal Individual Microbial Omics Networks
Creation and validation of LIMON - Longitudinal Individual Microbial Omics Networks
Alvernaz, S. A.; Penalver Bernabe, B.
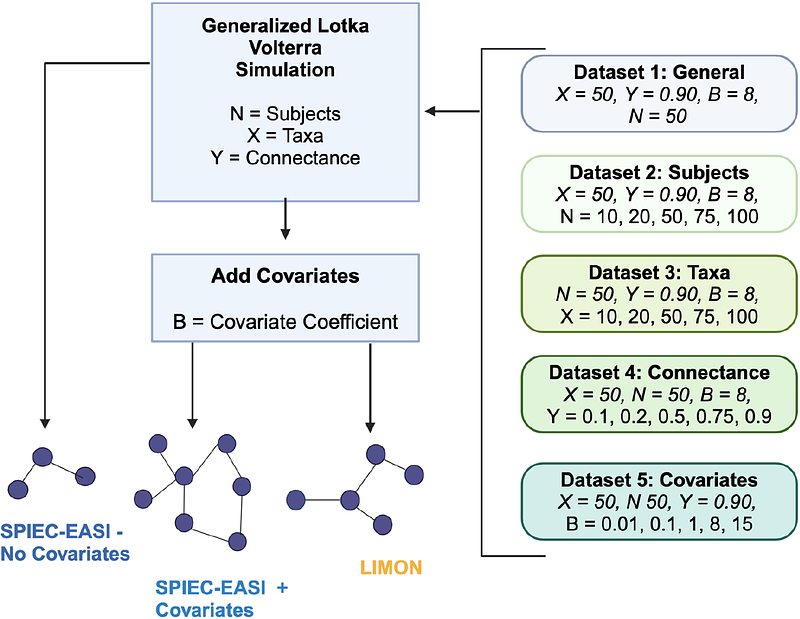
AbstractMicrobial communities are dynamic structures that continually adapt to their surrounding environment. Such communities play pivotal roles in countless ecosystems from environmental to human health. Perturbations of these community structures have been implicated in disease processes such as Crohns disease and cancer. Disturbances to existing ecosystems often occur over time, making it essential to have robust methods for detecting longitudinal alterations in microbial interactions as they develop. Existing methods for identifying temporal microbial community alterations have focused on abundance alterations in individual taxa, rather than relationships between the taxa, known as microbial interactions. Identifying these interactions overtime provides a fuller understanding of how the microbial ecosystem changes as a whole. To fill this gap, we have developed a pipeline that handles the complicated nature of repeated compositional count data, LIMON, Longitudinal Individual Microbial Omics Networks. This novel statistical approach addresses key challenges of modeling temporal and microbial data including overdispersion, zero inflated count data, compositionality, repeated measure design sample covariates over time, and identification of individualized or sample specific networks. This approach allows users to denoise covariate effects from their data, return networks per time point, identify interaction changes between each time point, and return individual networks and network characteristics per sample per time point. In doing so, LIMON provides a platform to identify the relationship between network interactions and sample features of interest over time. Here we show LIMON, in simulation studies, can accurately remove covariate effects, render sample specific networks, and better recover underlying network edges from covariate confounded data. Analysis of a longitudinal infant microbiome and diet dataset illustrates LIMONs novel utility to identify key microbial interactions related to diet type across time.