Collective unstructured interactions drive chromatin binding of transcription factors
Collective unstructured interactions drive chromatin binding of transcription factors
Abidi, A. A.; Graham, T. G. W.; Dailey, G. M.; Tjian, R.
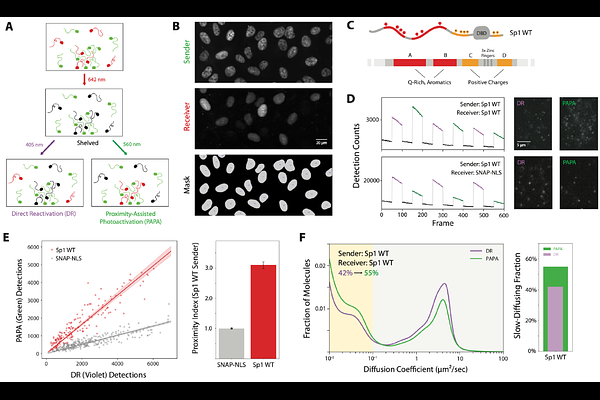
AbstractEukaryotic transcription factors (TFs) contain both structured DNA-binding domains (DBDs) and intrinsically disordered regions (IDRs). While the structures and sequence preferences of DBDs have been extensively characterized, the role of IDR-mediated interactions in chromatin binding and nuclear organization remains poorly understood, in part because these interactions have been difficult to measure in living cells. Here, we use a recently developed single-molecule technique, proximity-assisted photoactivation (PAPA), to investigate how IDRs influence TF associations with each other and with chromatin, focusing on the factors Sp1 and Klf1. We find that the number and patterning of aromatic and basic residues within IDRs govern both TF self-association and chromatin binding. Remarkably, the isolated DBD of Sp1 binds chromatin weakly and non-specifically. By contrast, the isolated IDR interacts transiently with chromatin-bound full-length Sp1, and these interactions become more stable when even minimal DNA-binding capacity is restored. Replacing Sp1's native DBD with domains from heterologous TFs with differing sequence preferences - or even with nonspecific bacterial DBDs -preserves IDR-mediated interactions and chromatin association. PAPA measurements also reveal extensive heterotypic interactions between Sp1 and other TFs. Together, these results establish PAPA as a powerful method for studying IDR interactions in their native context and suggest that IDRs participate in widespread cooperative associations scaffolded around chromatin. In contrast to classical models, we propose that TF specificity in vivo emerges from ensembles of weak, dynamic, and diverse interactions - not from the intrinsic sequence preferences of DBDs alone.