Neonatal infection with Helicobacter pylori affects stomach and colon microbiome composition and gene expression in mice
Neonatal infection with Helicobacter pylori affects stomach and colon microbiome composition and gene expression in mice
Graversen, K. B.; Bjarnov-Nicolau, B.; Klove, S.; Halajova, K.; Andersen, S. B.
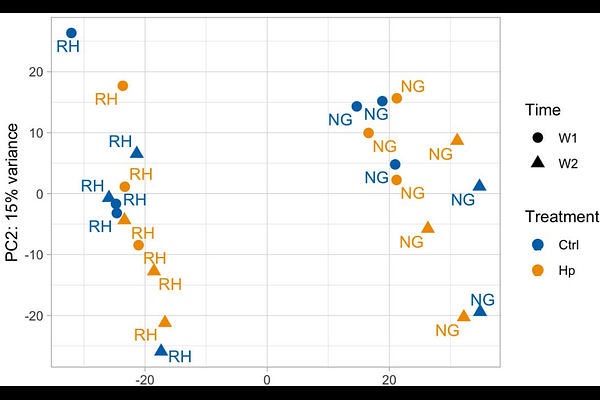
AbstractThe stomach bacterium Helicobacter pylori is estimated to infect half of the world\'s population, and the health implications are affected by the age at infection. Neonatal H. pylori infection of mice is a relevant model to investigate metabolic and immunological effects. We performed an explorative study at the dynamic first month of life, to compare the composition of the gastrointestinal tract microbiome and stomach gene expression of mice neonatally infected with H. pylori with that of uninfected mice. We found that H. pylori was present only in the stomach, and that H. pylori loads increase with age from one week after infection and onwards, especially after weaning. Stomach and colon microbiome composition was strikingly similar between sites at the same sampling time, but changed significantly over one week, with increased diversity at both sites. Despite that the relative abundance of H. pylori in the stomach was low and never exceeded 3%, the composition and alpha diversity of the gastrointestinal microbiome was significantly affected by infection. In a pathway enrichment analysis we found that stomach gene expression related to the extracellular matrix, muscle contraction, and metabolism was affected by infection. Expression of these key processes was, in infected mice, shifted away from that of control mice, towards that of all mice sampled the subsequent week, which we speculate represents accelerated development in infected mice.