Rhythmic variation in proteomics: challenges and opportunities for statistical power and biomarker identification
Rhythmic variation in proteomics: challenges and opportunities for statistical power and biomarker identification
Spick, M.; Isherwood, C. M.; Gethings, L.; Hassanin, H.; van der Veen, D. R.; Skene, D. J.; Johnston, J. D.
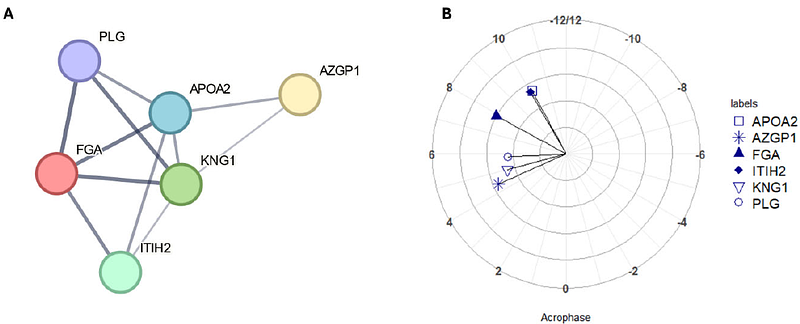
AbstractTime-of-day variation in the molecular profile of biofluids and tissues is a well-described phenomenon, but - especially for proteomics - is rarely considered in terms of the challenges this presents to reproducible biomarker identification. In this work we demonstrate these confounding issues using a small scale proteomics analysis of male participants in a constant routine protocol following an 8-day laboratory study, in which sleep-wake, light-dark and meal timings were controlled. We provide a case study analysis of circadian and ultradian rhythmicity in proteins in the complement and coagulation cascades, as well as apolipoproteins, and demonstrate that rhythmicity increases the risk of Type II errors due to the reduction in statistical power from increased variance. For the proteins analysed herein we show that to maintain statistical power if chronobiological variation is not controlled for, n should be increased (by between 9% and 20%); failure to do so would increase {beta}, the chance of Type II error, from a baseline value of 20% to between 22% and 28%. Conversely, controlling for rhythmic time-of-day variation in study design offers the opportunity to improve statistical power and reduce the chances of Type II errors. Indeed, control of time-of-day sampling is a more cost-effective strategy than increasing sample sizes. We recommend that best practice in proteomics study design should account for temporal variation as part of sampling strategy where possible. Where this is impractical, we recommend that additional variance from chronobiological effects be considered in power calculations, that time of sampling be reported as part of study metadata, and that researchers reference any previously identified rhythmicity in biomarkers and pathways of interest. These measures would mitigate against both false and missed discoveries, and improve reproducibility, especially in studies looking at biomarkers, pathways or conditions with a known chronobiological component.


