Assessment of machine-learning predictions for MED25 ACID domain interactions with transactivation domains
Assessment of machine-learning predictions for MED25 ACID domain interactions with transactivation domains
Monte, D.; Lens, Z.; Dewitte, F.; Villeret, V.; Verger, A.
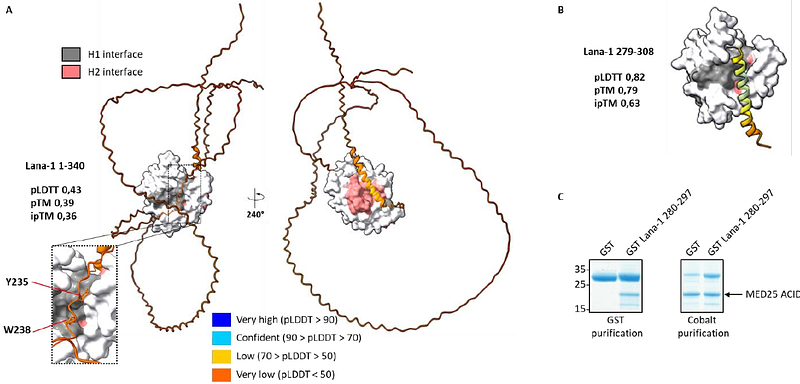
AbstractHuman Mediator complex subunit MED25 binds transactivation domains (TADs) present in various cellular and viral proteins using two binding interfaces found on opposite sides of its ACID domain, and referenced as H1 and H2. Here, we use and compare deep learning methods to characterize Human MED25-TADs interfaces and assess the predicted models to published experimental data. For the H1 interface, AlphaFold produces predictions with high reliability scores that agree well with experimental data, while the H2 interface predictions appear inconsistent, preventing reliable binding modes. Despite these limitations, we experimentally assess the validity of Lana-1 and IE62 MED25 interface predictions. AlphaFold predictions also suggest the existence of a unique hydrophobic pocket for Arabidopsis MED25 ACID domain.