A highly contiguous reference genome for the Alpine ibex (Capra ibex)
A highly contiguous reference genome for the Alpine ibex (Capra ibex)
Cilingir, F. G.; Landuzzi, F.; Brambilla, A.; Charrance, D.; Furia, F.; Trova, S.; Peracino, A.; Camenisch, G.; Waldvogel, D.; Howard-McCombe, J.; Castillo De Spelorzi, Y. C.; Henzen, E.; Bernagozzi, A.; Coppe, A.; Christille, J. M.; Vecchi, M.; Vozzi, D.; Cavalli, A.; Bassano, B.; Gustincich, S.; Croll, D.; Pandolfini, L.; Grossen, C.
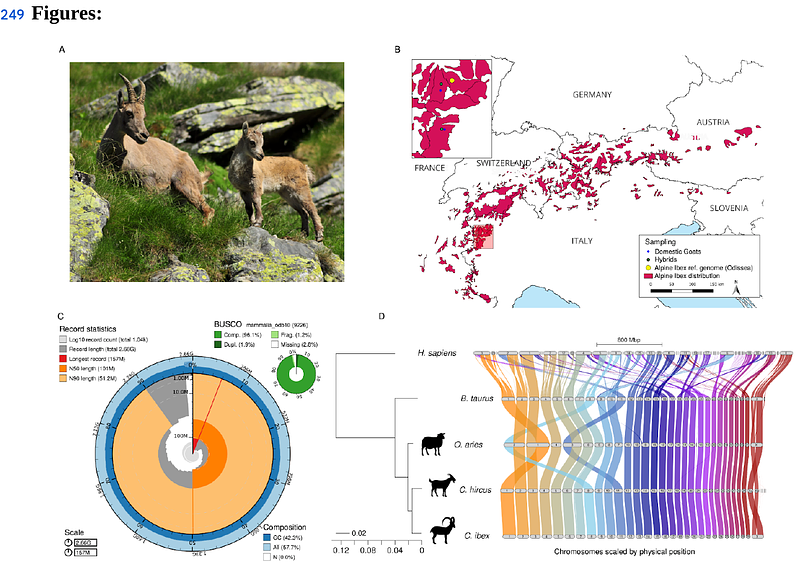
AbstractAnthropogenic hybridization, the unintentional hybridization with a non-native or domestic species in human-dominated environments, is a major concern for species conservation and a challenge for conservation management decisions. Genetically depleted species are expected to be particularly vulnerable to hybridization and introgression since hybridization can restore or introduce new adaptive genetic variation and alleviate the effects of inbreeding through hybrid vigour. However, defining the precise sets of deleterious or beneficial mutations resulting from anthropogenic hybridization is complex and limited by the quality of genomic resources. The Alpine ibex (Capra ibex), a species native to the Alps, faced near-extinction two centuries ago, but conservation programs have successfully restored its populations. Alpine ibex are known to hybridize with the domestic goat (Capra hircus) occasionally leading to hybrid swarm formation. Past introgression has been observed at immune-related genes and was suggested to have had an adaptive effect. Alpine ibex also carry deleterious mutation load from the reintroduction bottlenecks, which could be alleviated through admixture. Here, we produced a chromosome-level reference genome for Alpine ibex based on Oxford Nanopore sequencing coupled with high-throughput chromosome conformation capture. The highly contiguous assembly of 2.66 Gbp reveals 30 chromosomes and is augmented by a 98.8% complete RNAseq-assisted gene model prediction. The Alpine ibex genome presents overall a high degree of synteny compared to the domestic goat, with a number of structural variants spanning 38 Mb of insertion and deletions, as well as 49 Mb of inversions. We also identified structural variants near but not within the major histocompatibility complex (MHC), an immune-relevant gene complex, where previous studies found signals of introgression. The high degree of synteny between the Alpine ibex and domestic goat chromosomes likely facilitates recombination between haplotypes of the two species and is, therefore, in accordance with the observation of hybrid swarms. To determine the precise impact of recent admixture, we resequenced eight hybrid individuals sampled from two hybrid swarms in Northern Italy. Swarm individuals carried between 18-80% goat genome representing up to 3rd generation hybrids, including one F1 hybrid. The reference genome will facilitate quantifying maladaptive variation introduced from domestic goats and guide management efforts.


