ProkBERT PhaStyle: Accurate Phage Lifestyle Prediction with Pretrained Genomic Language Models
ProkBERT PhaStyle: Accurate Phage Lifestyle Prediction with Pretrained Genomic Language Models
Juhasz, J.; Bodnar, B.; Juhasz, J.; Ligeti-Nagy, N.; Pongor, S.; Ligeti, B.
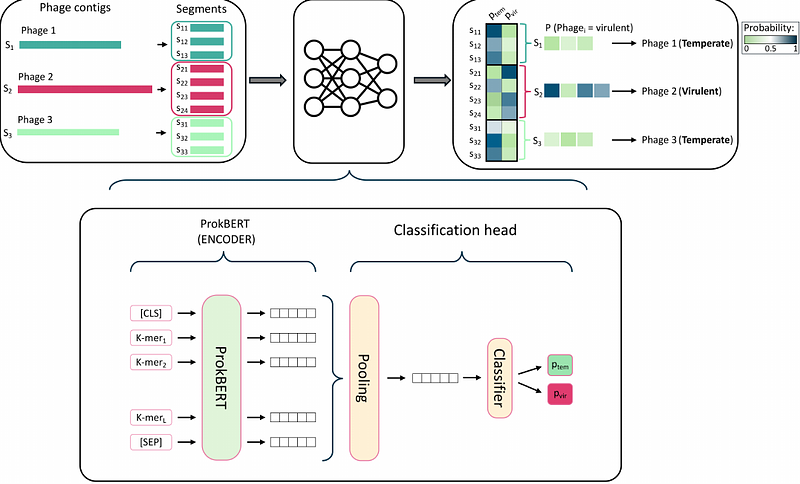
AbstractBackground: Phage lifestyle prediction, i.e. classifying phage sequences as virulent or temperate, is crucial in biomedical and ecological applications. Phage sequences from metagenome or metavirome assemblies are often fragmented, and the diversity of environmental phages is not well known. Current computational approaches often rely on database comparisons and machine learning algorithms that require significant effort and expertise to update. We propose using genomic language models for phage lifestyle classification, allowing efficient direct analysis from nucleotide sequences without the need for sophisticated preprocessing pipelines or manually curated databases. Methods: We trained three genomic language models (DNABERT-2, 117m parameters; Nucleotide Transformer, 50-500m parameters; and ProkBERT, 21-26m parameters) on datasets of short, fragmented sequences. These models were then compared with dedicated phage lifestyle prediction methods (PhaTYP, DeePhage, BACPHLIP) in terms of accuracy, prediction speed, and generalization capability. Results: ProkBERT PhaStyle consistently outperforms existing models in various challenging scenarios. It generalizes well for out-of-sample data, accurately classifes phages from extreme environments, and also demonstrates high inference speed. Despite having up to 20 times fewer parameters, it proved to be more performant than much larger genomic language models. Conclusions: Genomic language models offer a simple and computationally efficient alternative for solving complex classification tasks, such as phage lifestyle prediction. ProkBERT PhaStyle\'s simplicity, speed, and performance suggest its utility in various ecological and clinical applications.