Gene Loss DB: A curated database for gene loss in vertebrate species.
Gene Loss DB: A curated database for gene loss in vertebrate species.
Themudo, G. E.; Ruivo, R.; Valente, R.; Artilheiro, N.; Oliveira, D.; Amorim, I.; Pinto, B.; Castro, L. F. C.; Fernandes, S.; Lopes-Marques, M.
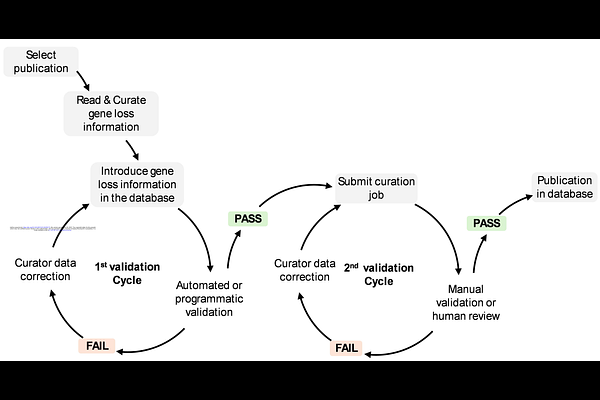
AbstractMolecular databases are essential resources for both experimental and computational biologists. The rapid increase in high-quality genome assemblies has led to a surge in publications describing secondary gene loss events associated with lineage-specific adaptations across diverse vertebrate groups. This growing volume of information underscores the urgent need for organized, searchable, and curated resources that facilitate data discovery, allow detection of broad evolutionary patterns, and support downstream analyses. Currently, no existing database compiles manually curated and validated information on published secondary gene loss events. Here, we introduce the Gene Loss Database (GLossDB), a platform designed to centralize and present this data in an easy-to-search and user-friendly format (https://geneloss.org/ ). GLossDB compiles gene loss events alongside supporting evidence, including the inferred mechanism of gene loss (exon deletion, gene deletion, loss of function mutation), the type of data used to support inactivation (genomic, transcriptomic, single/multiple individual sequence reads, synteny maps) and, when available, whether the event is shared across all lineages within a taxon. Each entry also includes a short excerpt from the original publication to provide context. This information is structured in the database to be searchable by species, gene, taxa, or by GO-terms linked to the gene in question. The initial release of GLossDB focuses on cetaceans, a lineage with numerous gene loss events linked to aquatic adaptations. This first collection comprises 1866 gene loss events identified across 57 cetacean species. In addition, the database includes 1321 gene loss events from other taxa, which were also reported in the same studies and collected simultaneously.