Characterizing antibodies binding to the same epitope reveals limited contribution by heavy chain CDR3 sequences relative to other CDRs
Characterizing antibodies binding to the same epitope reveals limited contribution by heavy chain CDR3 sequences relative to other CDRs
Mahita, J.; Kim, H.; De Almeida Mendes, M.; Richardson, E.; Greenbaum, J. A.; Sette, A.; Nielsen, M.; Peters, B.
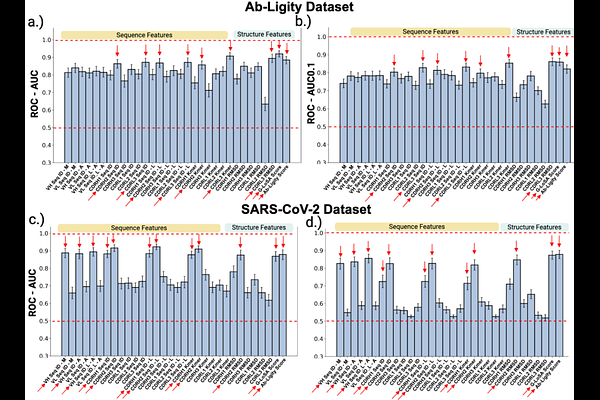
AbstractThe antigen specificity of antibodies is defined mainly by their highly-variable complementarity-determining regions (CDRs), of which the CDR3 is considered to be the most important. Similar antibodies are expected to bind the same epitope on an antigen, but how this similarity should be quantified is unclear. Here, we utilized a large body of publicly available data to identify features that are most informative in predicting if two antibodies are likely to bind to the same epitope. We examined features such as pairwise sequence identities, CDR structural similarity, and geometrical similarity of antibody paratopes. We found that, for antibodies with high overall sequence similarity, the CDR sequence identity alone is sufficient to predict if a pair of antibodies binds to the same epitope. Strikingly, the sequence identity of CDR1 and CDR2 had as much or more predictive power than CDR3, which is commonly thought of as the main determinant of antibody specificity. In addition, for antibody pairs with lower overall sequence identity, including structural information led to significant improvements in predictive performance. Guided by these results, we developed BCRMatch (https://github.com/IEDB/BCRMatch) which uses an ensemble machine-learning approach to predict if pairs of antibodies target the same epitope based on antibody sequence alone. This can be applied to identify potential targets of an antibody of interest by comparing it to antibodies with known specificities.