The Mechanism of Histone Ubiquitylation by the ASB9-CUL5 Ubiquitin Ligase
The Mechanism of Histone Ubiquitylation by the ASB9-CUL5 Ubiquitin Ligase
Komives, E. A.; Lee, N. H.; Alipranti, F. X.; Li, H.; Lin, C. P.
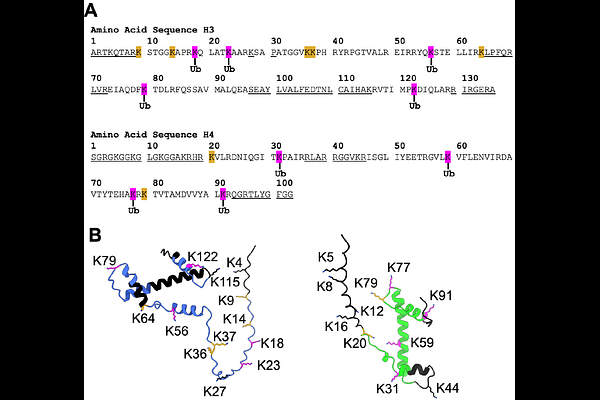
AbstractThe E3 ligase substrate receptor ankyrin and SOCS box protein 9 (ASB9) was shown to bind over 10 different proteins including metabolic enzymes such as creatine kinase, filament proteins such as vimentin, and histones. In previous work, we characterized the ASB9-Cullin 5 E3 ligase (ASB9-CRL) ubiquitylation of creatine kinase and showed that ubiquitylation required the ring-between-ring ligase, ARIH2. Here we characterize the ASB9-CRL ubiquitylation of histones and show that histones H3 and H4 are polyubiquitylated by the ASB9-CRL whereas histones H2A and H2B are not. Many, but not all lysines in the histones are ubiquitylated suggesting some substrate specificity. Binding experiments show that the ligase-histone interaction is highly electrostatic and the neddylated ASB9-CRL binds with highest affinity. Only free histones are ubiquitylated. When the histones are in nucleosomes or in complex with the chaperone Asf1, they are not ubiquitylated. Only K48 and K63 polyubiquitin chains were observed, suggesting that the ubiquitylation probably drives protein degradation. The presence of ASB9 in specific cell types correlates with situations in which free histones H3 and H4 need to be degraded. In this work, we demonstrate that the ASB9-CRL is the ligase that facilitates degradation of histones H3 and H4. In addition, this work represents the first example of Cullin 5 mediated ubiquitylation that does not require an additional ring-between-ring ligase.