PepMSND: Integrating Multi-level Feature Engineering and Comprehensive Databases to Enhance in vivo/in vitro Peptide Blood Stability Prediction
PepMSND: Integrating Multi-level Feature Engineering and Comprehensive Databases to Enhance in vivo/in vitro Peptide Blood Stability Prediction
Haomeng, H.; Zhang, C.; Zhenyu, X.; Duan, H.
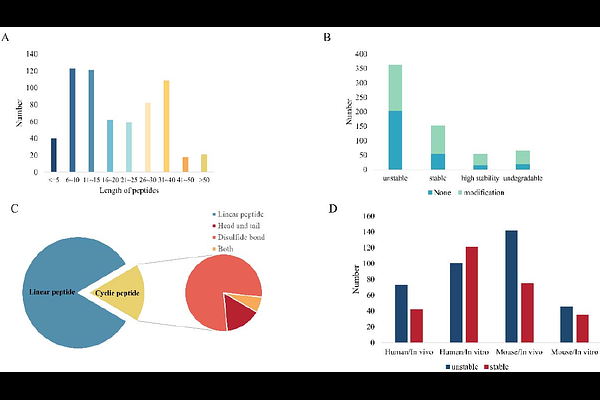
AbstractDeep learning technology has revolutionized the field of peptides, but key questions such as how to predict the blood stability of peptides remain. While such a task can be accomplished by experiments, it requires much time and cost. Here, to address this challenge, we collect extensive experimental data on peptide stability in blood from public databases and literature and construct a database of peptide blood stability that includes 635 samples. Based on this database, we develop a novel model called PepMSND, integrating KAN, Transformer, GAT and SE(3)-Transformer to make multi-level feature engineering to make peptide stability prediction. Our model can achieve the ACC of 0.8672 and the AUC of 0.9118 on average and outperforms the baseline models. This work can facilitate the development of novel peptides with strong stability, which is crucial for their therapeutic use in clinical applications.