Comparison and benchmark of deep learning methods for non-coding RNA classification
Comparison and benchmark of deep learning methods for non-coding RNA classification
Creux, C.; Zehraoui, F.; Radvanyi, F.; Tahi, F.
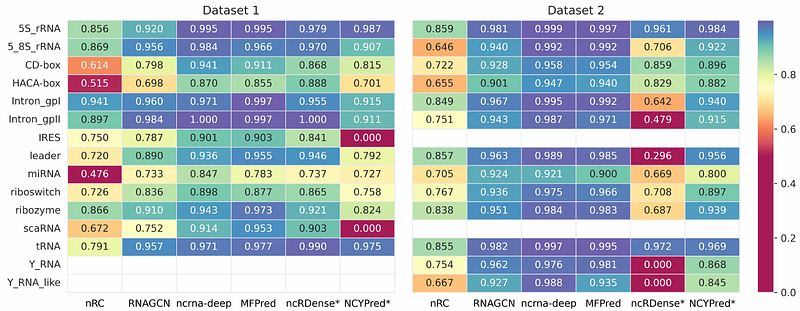
AbstractThe grouping of non-coding RNAs into functional classes started in the 1950s with housekeeping RNAs. Since, multiple additional classes were described. The involvement of non-coding RNAs in biological processes and diseases has made their characterization crucial, creating a need for computational methods that can classify large sets of non-coding RNAs. In recent years, the success of deep learning in various domains led to its successful application to non-coding RNA classification. Multiple novel architectures have been developed, but these advancements are not covered by current literature reviews. We propose a comparison of the different approaches and of ncRNA datasets proposed in the state-of-the-art. Then, we perform experiments to fairly evaluate the performance of various tools for non-coding RNA classification. With regard to these results, we assess the relevance of the different architectural choices and provide recommendations to consider in future methods.