CRISPR/Cas9-induced double-strand breaks in huntingtin locus lead to CAG repeat contraction through the extensive DNA end resection and homology-mediated repair
CRISPR/Cas9-induced double-strand breaks in huntingtin locus lead to CAG repeat contraction through the extensive DNA end resection and homology-mediated repair
Sledzinski, P.; Nowaczyk, M.; Karwacka, M. I.; Olejniczak, M.
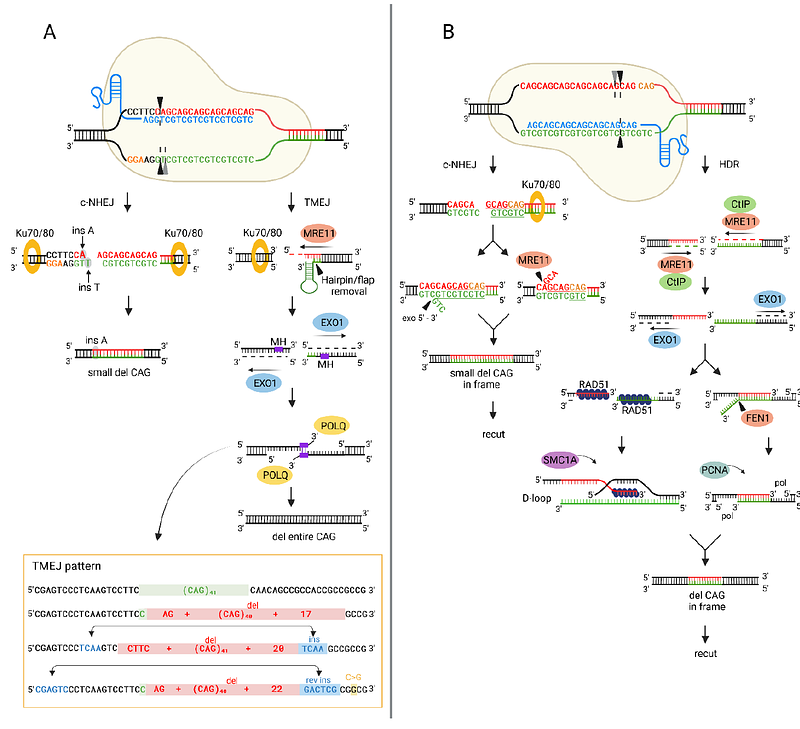
AbstractExpansion of the CAG/CTG repeats in functionally unrelated genes is a causative factor in many inherited neurodegenerative disorders, including Huntington\'s disease (HD), spinocerebellar ataxias (SCAs) and myotonic dystrophy type 1 (DM1). Despite many years of research, the mechanism responsible for repeat instability is unknown, and recent findings indicate the key role of DNA repair in this process. The repair of DSBs induced by genome editing tools results in the shortening of long CAG repeats in yeast models. Understanding this mechanism is the first step to developing a therapeutic strategy based on controlled shortening of repeats. The aim of this study was to characterize Cas9-induced DSB repair products in the endogenous HTT locus in human cells and to identify factors affecting the formation of specific types of sequences. The location of the cleavage site and the surrounding sequence influence the outcome of DNA repair. DSBs within CAG repeats result in shortening of the repeats in frame in ~90% of products. The mechanism of this contraction involves MRE11-CTIP and RAD51 activity and extensive DNA end resection reaching ~5000 bp. We demonstrated that a DSB located upstream of CAG repeats induces polymerase theta-mediated end joining, resulting in deletion of the entire CAG tract. Furthermore, using unbiased proteomic analysis, we identified novel factors that may be involved in CAG sequence repair.