DIGEST: An online tool for designing of multiple reaction monitoring assays
DIGEST: An online tool for designing of multiple reaction monitoring assays
gautam, p.; Singh, P.; Mrinal, ; Bhaskar, A.; Sacher, S.; Dagar, Y.; Basak, T.; Sengupta, S.; Ray, A.
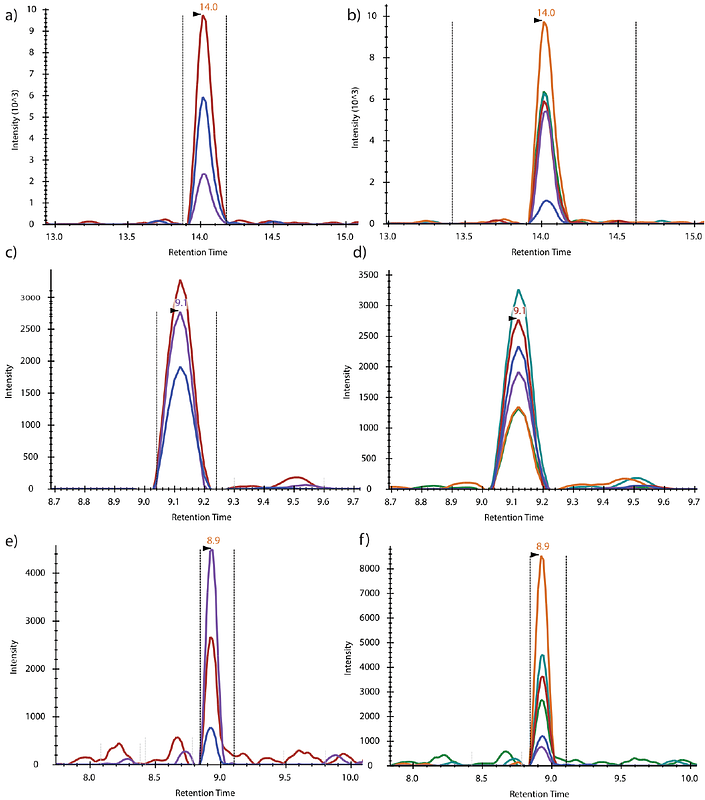
AbstractTargeted proteomics using multiple reaction monitoring (MRM) assays enables fast and sensitive detection of a preselected set of target peptides. This technique utilizes the specificity of precursors to product transitions for quantitative analysis of multiple proteins in a single sample. The success of an MRM experiment depends on the selection of transitions however, given the existing resources, accurately predicting signal intensity of peptides and their fragmentation patterns ab initio is challenging task. We present an alternative for rapid design of MRM transitions for proteomics research: DIGEST. Our method predicts the b and y ions with +1 and +2 charge produced in a collision cell of a mass spectrometer from peptides of multiple proteotypically digested proteins. Additionally, by using the existing knowledge of the fundamental rules for designing transitions, the tool provides optimal MRM transitions, negating the need to undertake prior discovery MS studies. We demonstrate that our algorithm is directed toward the selection of MRM precursor and product-ions pairs, and can avoid the pitfalls of interference due to cross-contamination of samples by selecting ion combinations that uniquely map to target peptides. Comparison with SRMAtlas showed that DIGEST successfully predicted the peptide and product-ion pairs in the majority of cases. We believe that DIGEST will facilitate rapid design of MRM assays with increased specificity, reducing the overall time required to design an MRM assay for routine mass-spectrometry. DIGEST is available as a web-based tool at https://digest.raylab.iiitd.edu.in/