Stochastic Phylogenetic Models of Shape
Stochastic Phylogenetic Models of Shape
Stroustrup, S.; Pedersen, M. A.; van der Meulen, F.; Sommer, S.; Nielsen, R.
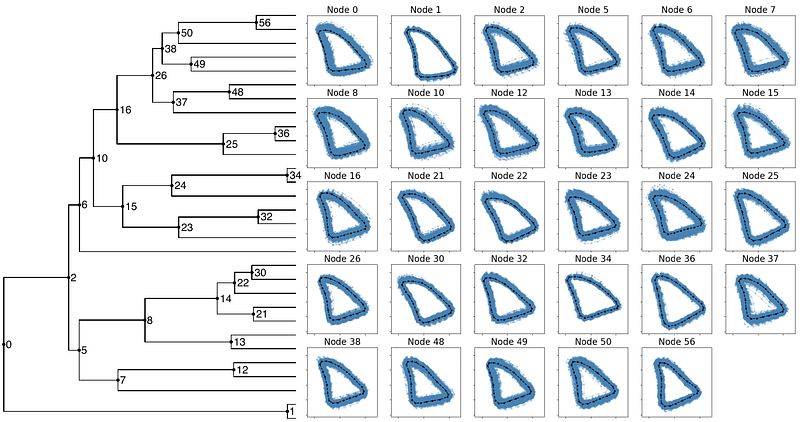
AbstractPhylogenetic modeling of morphological shape in two or three dimensions is one of the most challenging statistical problems in evolutionary biology. As shape data are inherently correlated and non-linear, most naive methods for phylogenetic analysis of morphological shape fail to capture the biological realities of evolving shapes. In this study we propose a novel framework for evolutionary analysis of morphological shape which facilitates stochastic character mapping on landmark shapes. The framework is based on recent advances in mathematical shape analysis and models the evolution of shape as a diffusion process that accounts for the evolutionary correlation between nearby landmarks. It uses a Metropolis-Hastings Markov Chain Monte Carlo sampling scheme for inferring ancestral shape and parameters of the model. We evaluate the new inference algorithm using simulations and show that the method leads to improved estimates of the shape at the root and well-calibrated credible sets of shapes at internal nodes. To illustrate the method, we also apply it to a previously published data set of butterfly wing images.