Iterative deep learning-design of human enhancers exploits condensed sequence grammar to achieve cell type-specificity
Iterative deep learning-design of human enhancers exploits condensed sequence grammar to achieve cell type-specificity
Yin, C. H.; Castillo Hair, S.; Woo Byeon, G.; Bromley, P.; Meuleman, W.; Seelig, G.
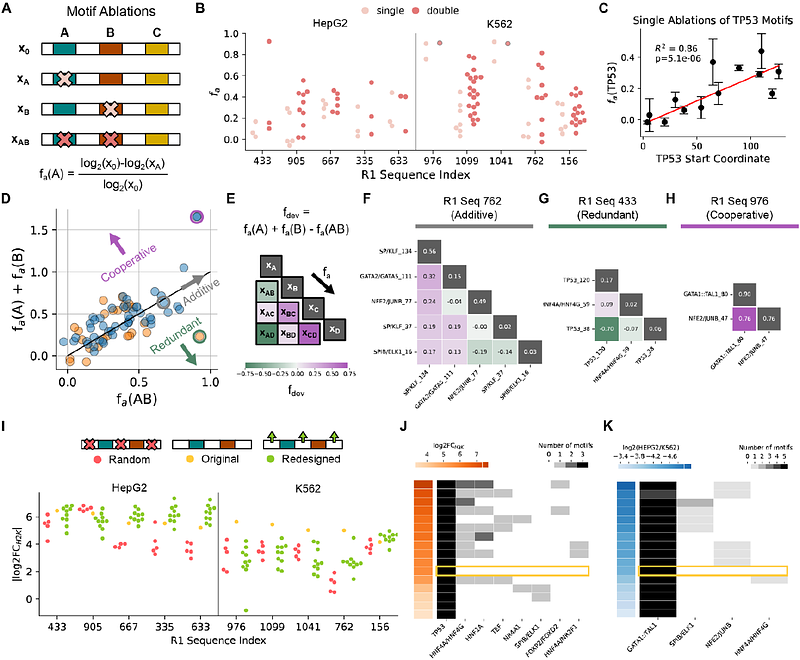
AbstractAn important and largely unsolved problem in synthetic biology is how to target gene expression to specific cell types. Here, we apply iterative deep learning to design synthetic enhancers with strong differential activity between two human cell lines. We initially train models on published datasets of enhancer activity and chromatin accessibility and use them to guide the design of synthetic enhancers that maximize predicted specificity. We experimentally validate these sequences, use the measurements to re-optimize the predictor, and design a second generation of enhancers with improved specificity. Our design methods embed relevant transcription factor binding site (TFBS) motifs with higher frequencies than comparable endogenous enhancers while using a more selective motif vocabulary, and we show that enhancer activity is correlated with transcription factor expression at the single cell level. Finally, we characterize causal features of top enhancers via perturbation experiments and show enhancers as short as 50bp can maintain specificity.


