Creation of de novo cryptic splicing for ALS/FTD precision medicine
Creation of de novo cryptic splicing for ALS/FTD precision medicine
Wilkins, O. G.; Chien, M. Z. Y. J.; Wlaschin, J. J.; Pisliakova, M.; Thompson, D.; Digby, H.; Simkin, R. L.; Diaz, J. A.; Mehta, P. R.; Keuss, M. J.; Zanovello, M.; Brown, A.-L.; Harley, P.; Darbey, A.; Karda, R.; Fisher, E. M. C.; Cunningham, T. J.; Le Pichon, C. E.; Ule, J.; Fratta, P.
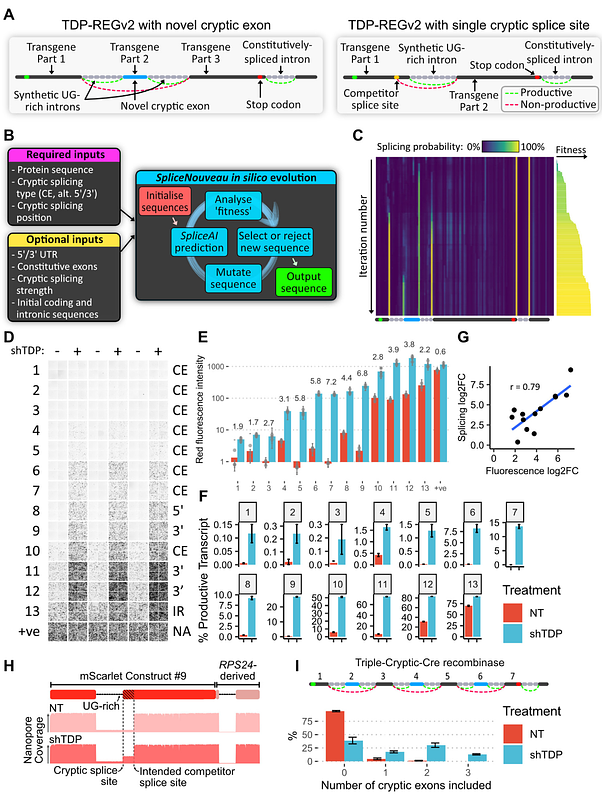
AbstractA system enabling the expression of therapeutic proteins specifically in diseased cells would be transformative, providing greatly increased safety and the possibility of pre-emptive treatment. Here we describe \'TDP-REG\', a precision medicine approach primarily for amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), which exploits the cryptic splicing events that occur in cells with TDP-43 loss-of-function (TDP-LOF) in order to drive expression specifically in diseased cells. In addition to modifying existing cryptic exons for this purpose, we develop a deep-learning-powered algorithm for generating customisable cryptic splicing events, which can be embedded within virtually any coding sequence. By placing part of a coding sequence within a novel cryptic exon, we tightly couple protein expression to TDP-LOF. Protein expression is activated by TDP-LOF in vitro and in vivo, including TDP-LOF induced by cytoplasmic TDP-43 aggregation. In addition to generating a variety of fluorescent and luminescent reporters, we use this system to perform TDP-LOF-dependent genomic prime editing to ablate the UNC13A cryptic donor splice site. Furthermore, we design a panel of tightly gated, autoregulating vectors encoding a TDP-43/Raver1 fusion protein, which rescue key pathological cryptic splicing events. In summary, we combine deep-learning and rational design to create sophisticated splicing sensors, resulting in a platform that provides far safer therapeutics for neurodegeneration, potentially even enabling preemptive treatment of at-risk individuals.