Using domain insertion to create sulfite reductases that present chemical-dependent activities
Using domain insertion to create sulfite reductases that present chemical-dependent activities
Windham, E.; Myerscough, D.; Schwartz, S.; Carpenter, M.; Ajo-Franklin, C.; Silberg, J.
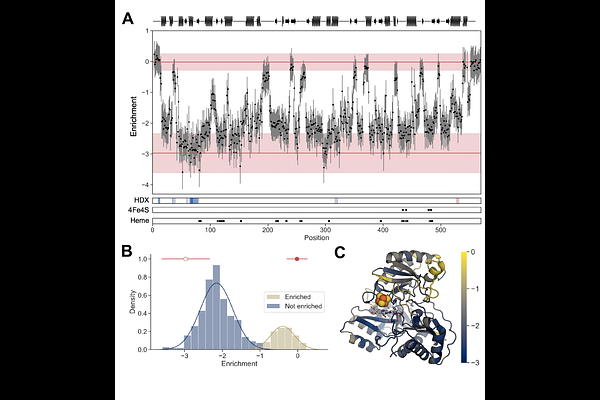
AbstractDomain insertion can be used to create oxidoreductase switches whose charge transfer is dependent upon analyte binding. To date, most domain insertion studies have targeted relatively small proteins of known structure, so it remains unclear how to effectively use this protein engineering approach with large oligomeric oxidoreductases that require dynamic conformational changes for catalysis. To address this question, we studied the effect of domain insertion on the function of NADPH-dependent sulfite reductase (SiR) from Escherichia coli, a dodecameric protein containing eight hemoprotein subunits and four flavoprotein subunits. SiR mutational tolerance was first mapped using systematic peptide insertion, and a subset of variants retaining activity were subjected to domain insertion. When a ligand-binding domain was inserted at locations tolerant to peptide insertion, including sites proximal and distal from the intersubunit interfaces, more than half of the resulting variants presented cellular activity that is enhanced by an endocrine disruptor. This ligand-dependent synthesis of a redox-active metabolite could be monitored electrochemically from cells, illustrating how a single protein complex can be used to convert chemical information in the environment into a metabolite within cells that diffuses across the cell membrane and can be detected electrochemically.