Concurrent detection of chemically modified bases in yeast mitochondrial tRNAs by Nanopore direct RNA sequencing
Concurrent detection of chemically modified bases in yeast mitochondrial tRNAs by Nanopore direct RNA sequencing
Reinsch, J. L.; Garcia, D. M.
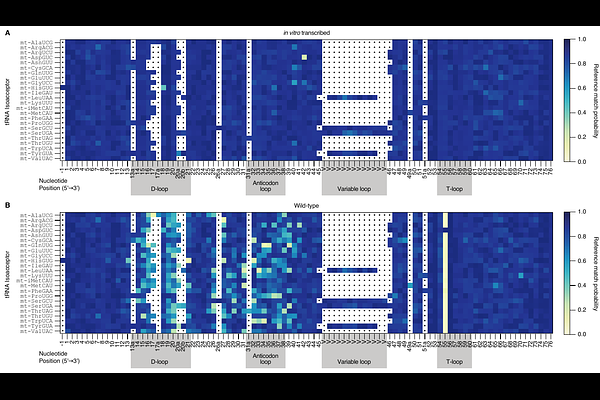
AbstractMutations in tRNA-modifying enzymes are associated with severe mitochondrial disease in humans. Saccharomyces cerevisiae has been used to model the impact of these mutations due to enzyme conservation and the ease of phenotyping mitochondrial defects in yeast. Yet the mature sequences of all its mitochondrially-encoded tRNAs (mt-tRNAs), including the positions of chemically modified bases, have not been determined in their entirety. We used Nanopore direct RNA sequencing (DRS) of mitochondrially-enriched RNA combined with enzyme knockouts to map base modifications across 24 mt-tRNA isoacceptors in yeast. Analysis of mt-tRNAs from wild-type yeast revealed base miscalls that were coincident with known modification sites at annotated positions, as well as at previously undescribed sites. Comparison to yeast lacking the PUS4 gene, which encodes a conserved pseudouridine synthase, demonstrated that 23 of 24 mt-tRNAs are pseudouridylated by Pus4 to form {Psi}55 in the T-loop. We also mapped the uridines modified by Pus2, catalyzing {Psi}27 and/or {Psi}28 on several mt-tRNAs, a function matching its paralog Pus1 that modifies cytosolic tRNAs at the same positions. In yeast lacking the PUS2 gene, some mt-tRNAs also showed changes at other positions inconsistent with being Pus2-pseudouridylated sites. These include U34 that is modified to cmnm5s2U34 in the anticodon of mt-tRNALys(UUU), and A21 in mt-tRNAHis(GUG) that is not previously known to be modified. Sequencing of a strain lacking the DUS2 dihydrouridine synthase gene also permitted determination of the dihydrouridine sites catalyzed by Dus2. Loss of this D20 modification also led to pleiotropic effects on other putative modification sites, those presumed catalyzed by Dus1 and Trm1, in many mt-tRNAs. In summary these data provide a comprehensive analysis of S. cerevisiae mt-tRNA sequences, including modifications. They also suggest previously unknown RNA modification <q>circuits</q> in mt-tRNAs, in which loss of Pus2-catalyzed {Psi}27/{Psi}28 and Dus2-catalyzed D20 causes increases or decreases in other modifications. Changes of multiple different modifications from loss of the activity of a single enzyme has implications for how mutations in these genes may cause pleiotropic effects on tRNA structure and translation.