Accurate Protein-Protein Interactions Modeling through Physics-informed Geometric Invariant Learning
Accurate Protein-Protein Interactions Modeling through Physics-informed Geometric Invariant Learning
Rao, J.; Liu, D.; Zhou, X.; Yuan, Q.; Wei, W.; Lu, W.; Zhang, J.; Rong, Y.; Yang, Y.; Zheng, S.
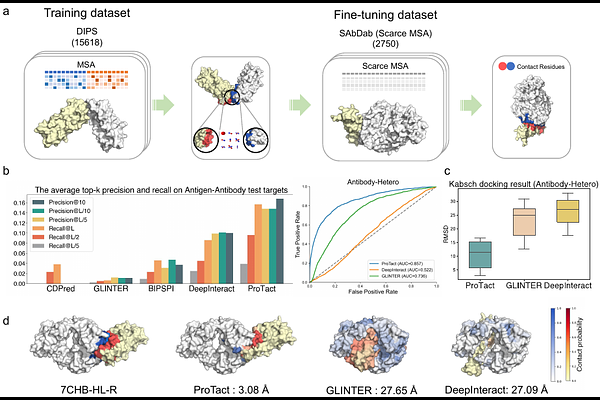
AbstractAlphaFold has set a new standard for predicting protein structures from primary sequences; however, it faces challenges with protein complexes across species, engineered proteins, and antigen-antibody, where co-evolutionary signals may be sparse or missing. Herein, we present ProTact, a SE(3)-invariant geometric graph neural network that integrates physics-informed geometric complementarity and trigonometric constraints as robust inductive biases to enhance protein-protein contact predictions. ProTact is applicable to both experimental and predicted monomer structures and utilizes a modulated key point matching algorithm to approximate accurate docking poses. Experimental evaluations demonstrate that ProTact consistently outperforms state-of-the-art sequence-based and structure-based methods on benchmark datasets, achieving notable relative improvements of 31.63% in Precision@10 for CASP 13 and 14 targets, and 31.94% for DIPS-Plus datasets. Moreover, when combined with AlphaFold3 as re-scoring functions, ProTact surpasses its default confidence scores, offering over 30.48% improvements in the low-MSA contexts. We anticipate that the proposed framework will advance our understanding of protein interactions, functions, and design.