Graphical and Interactive Spatial Proteomics Image Analysis Workflow
Graphical and Interactive Spatial Proteomics Image Analysis Workflow
Singh, P.; Wright, J. H.; Smythe, K. S.; Fukuda, B. N.; Hung, L.-H.; Yeung, C. C.; Yeung, K. Y.
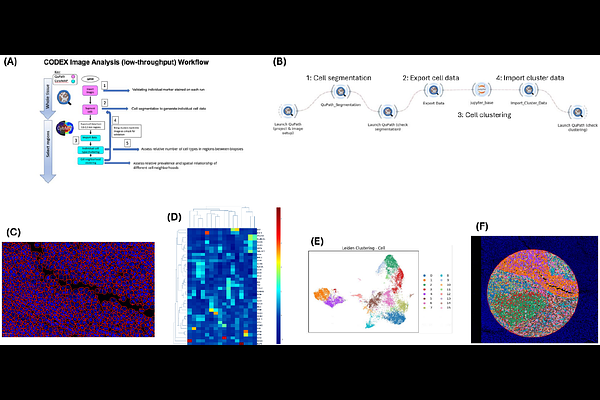
AbstractSpatial proteomics provides a spatially resolved view of protein expression and localization within cells and tissues by mapping the location and abundance of proteins. There is a need for containerized end-to-end imaging workflows for spatial proteomic analysis that are flexible, high-throughput, and support graphical and interactive visualizations. We present a modular and interactive spatial proteomics imaging workflow that empowers biomedical researchers to reproducibly execute and customize complex analyses. Our workflow consists of cell segmentation, unsupervised clustering, validation of clusters on the image, and cell type clustering results visualization. Users can utilize a form-based graphical interface to execute and customize multi-step workflows with a single click or interactively adjust image processing steps within the workflow, apply workflows to various datasets, and modify input parameters as needed. We illustrated the functionality of our workflow using a cancer imaging dataset consisting of a tissue microarray (TMA) stained by high-plex immunohistochemistry. This TMA contained a variety of cancer and tissue cell types to assess the broad applicability of this workflow to different biopsy types.