MolUNet++: Adaptive-grained Explicit Substructure and Interaction Aware Molecular Representation Learning
MolUNet++: Adaptive-grained Explicit Substructure and Interaction Aware Molecular Representation Learning
XU, F.; Yang, Z.; Su, W.; Wang, L.; Meng, D.; Long, J.
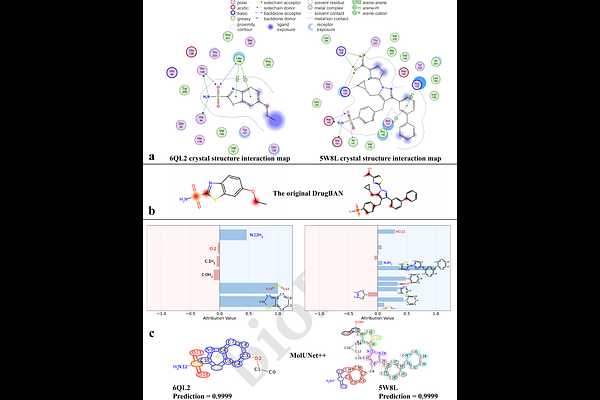
AbstractMolecular representation learning is a critical task in AI-driven drug development. While graph neural networks (GNNs) have demonstrated strong performance and gained widespread adoption in this field, efficiently extracting and explicitly analyzing functional groups remains a challenge. To address this issue, we propose MolUNet++, a novel model that employs Molecular Edge Shrinkage Pooling (MESPool) for hierarchical substructure extraction, utilizes a Nested UNet framework for multi-granularity feature integration, and incorporates a substructure masking explainer for quantitative fragment analysis. We evaluated MolUNet++ on tasks including molecular property prediction, drug-drug interaction (DDI) prediction, and drug-target interaction (DTI) prediction. Experimental results demonstrate that MolUNet++ not only outperforms traditional GNN models in predictive performance but also exhibits explicit, intuitive, and chemically logical interpretability. This capability provides valuable insights and tools for researchers in drug design and optimization.