Bursty Gene Expression in Single Cells and Expanding Populations: A Discrete Approach
Bursty Gene Expression in Single Cells and Expanding Populations: A Discrete Approach
Poljovka, J.; Zabaikina, I.; Bokes, P.; Singh, A.
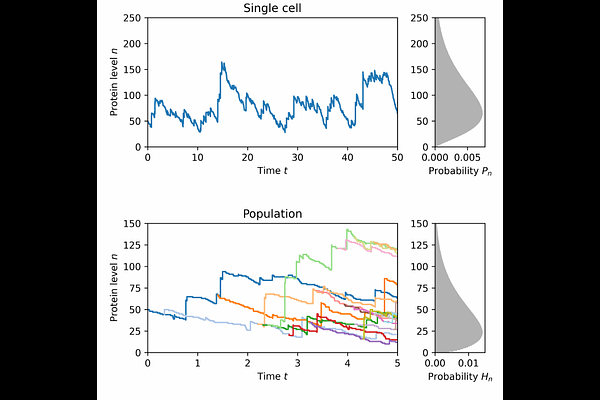
AbstractBursty protein production is a key source of gene expression noise. In this study, we analyze a Markovian model of cellular protein dynamics, where proteins are produced in geometrically distributed bursts. Extending this model, we incorporate a feedback mechanism by assuming that higher protein levels reduce both cell growth and protein decay rates. We study both single-cell dynamics and an expanding cell population. Without feedback, the protein level follows a negative binomial distribution with the same parameters in both cases. With feedback, however, single-cell and population-level distributions differ, each expressible as a mixture of two negative binomial distributions with framework-dependent parameters. Using numerical integration of the master and population balance equations, we calculate the time-dependent distributions in both settings. This work extends previous continuous models and provides new insights into how population expansion influences intrinsic cellular heterogeneity.