Climate associated natural selection in the human mitochondrial genome
Climate associated natural selection in the human mitochondrial genome
Grover-Thomas, F.; van Dorp, L.; Balloux, F.; Andres, A. M.; Camus, M. F.
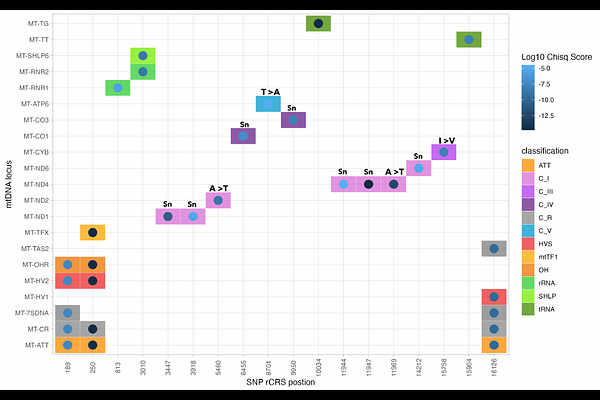
AbstractMitochondria are essential for cellular energy production and biosynthesis, thermogenesis, and cell signalling, and thus help coordinate physiological responses to changing environments. Humans (Homo sapiens) have adapted to cope with a wide range of climatic conditions, however the role of the mitochondrial genome (mtDNA) in mediating this process remains poorly understood. Here we curated a dataset of 23,790 publicly available full human mitochondrial genomes, an approximate 40-fold increase on earlier studies, paired with modern climate and reconstructed paleoclimate variables. Using a Generalised Linear Model approach, we identify 18 independent candidate variants significantly associated with climatic conditions, suggesting local adaptation in human mitochondrial genomes. Candidate variants are distributed across multiple loci in regulatory, tRNA, rRNA and protein-coding regions - including prominently in ND2 and ND4 complex I subunits. Specific variants are predicted to impact mtDNA transcription, ribosome or protein structure, and multiple have been associated with disease pathologies. We further show that candidate variant genotype distributions are each best modelled by different paleo-bioclimatic variables, consistent with environmental stressors linked to our measured variables exerting subtly distinct selective effects. These stressors may reflect dietary changes or different thermogenic demands at lower temperatures. Our results provide genetic evidence to support the accumulating body of work from functional studies that mitochondria can modulate adaptation to diverse environments. This work underscores the importance of mtDNA in evolutionary biology and its relevance for understanding both disease and physiological variation in global populations.