Thermal proteome profiling identifies glucose binding proteins involved in metabolic disease
Thermal proteome profiling identifies glucose binding proteins involved in metabolic disease
Ferguson, I. D.; Meservey, L. M.; Tien, V.; Miao, W.; Lopez-Pajares, V.; Porter, D. F.; Khavari, P.
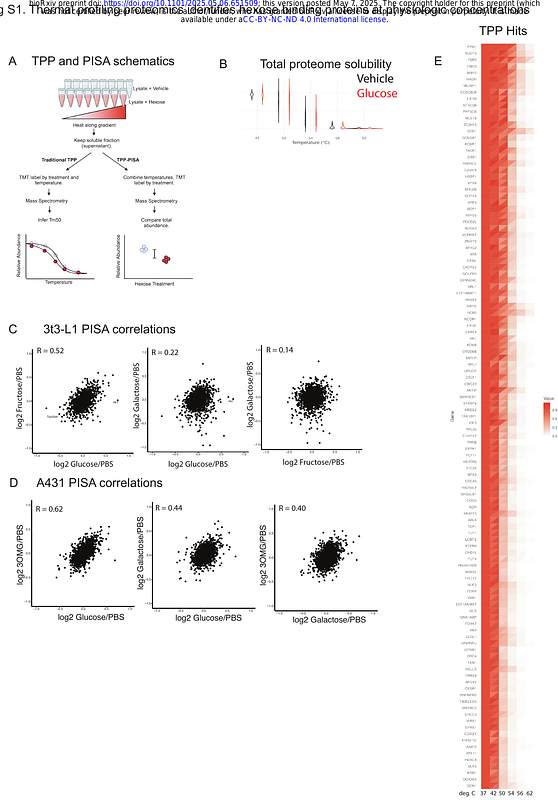
AbstractRecent work has shown that glucose can modulate protein multimerization and function independent of metabolism through direct protein binding. We sought to build on this work by adapting thermal proteome profiling screens to uncover novel glucose binders involved in metabolism. We found that TSC22D4, an intrinsically-disordered, leucine-zipper protein previously linked to insulin resistance and liver fat accumulation, exhibits altered thermal stability in the presence of glucose. Furthermore, glucose decreased TSC22D4 DNA binding and altered TSC22D4 protein-protein association. Employing UV-crosslinking mass spectrometry, we found glucose binds near the C-terminal leucine zipper domain. Mutating isoleucine 322 to tryptophan (I322W) abolishes glucose binding. Crosslinking-MS and chemo-proteomic experiments suggest that glucose increases accessibility of the leucine zipper region, resulting in intra-protein contacts between C-terminal zipper domain and N-terminal intrinsically-disordered domain. TSC22D4 is essential for adipogenesis in the 3T3-L1 model system, and glucose can increase nuclear localization of TSC22D4 and remodel its protein-protein interactions. Finally, we further extend this work by validating site-specific glucose binding for another PISA target with genetic linkage to metabolic disease.