SpatialCells: Automated Profiling of Tumor Microenvironments with Spatially Resolved Multiplexed Single-Cell Data
SpatialCells: Automated Profiling of Tumor Microenvironments with Spatially Resolved Multiplexed Single-Cell Data
Wan, G.; Maliga, Z.; Yan, B.; Vallius, T.; Shi, Y.; Khattab, S.; Chang, C.; Nirmal, A. J.; Yu, K.-H.; Liu, D.; Lian, C. G.; DeSimone, M. S.; Sorger, P. K.; Semenov, Y. R.
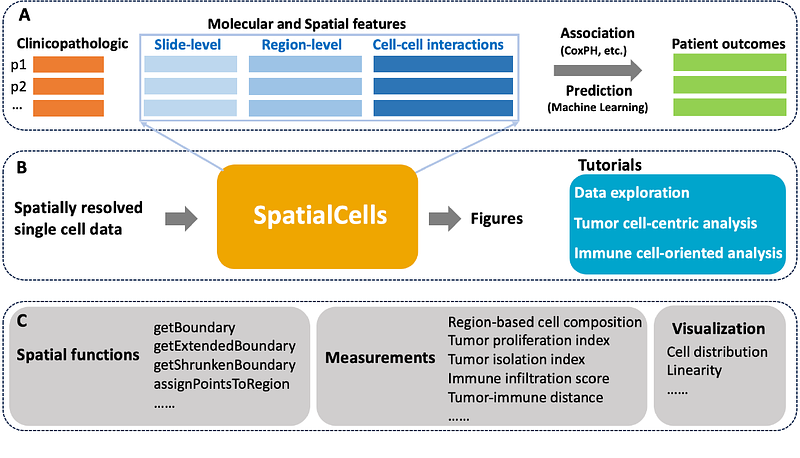
AbstractBackground: Cancer is a complex cellular ecosystem where malignant cells coexist and interact with immune, stromal, and other cells within the tumor microenvironment. Recent technological advancements in spatially resolved multiplexed imaging at single-cell resolution have led to the generation of large-scale and high-dimensional datasets from biological specimens. This underscores the necessity for automated methodologies that can effectively characterize the molecular, cellular, and spatial properties of tumor microenvironments for various malignancies. Results: This study introduces SpatialCells, an open-source software package designed for region-based exploratory analysis and comprehensive characterization of tumor microenvironments using multiplexed single-cell data. Conclusions: SpatialCells efficiently streamlines the automated extraction of features from multiplexed single-cell data and can process samples containing millions of cells. Thus, SpatialCells facilitates subsequent association analyses and machine learning predictions, making it an essential tool in advancing our understanding of tumor growth, invasion, and metastasis.