APPLE-MS: A affinity purification-mass spectrometry method assisted by PafA-mediated proximity labeling
APPLE-MS: A affinity purification-mass spectrometry method assisted by PafA-mediated proximity labeling
Luo, S.; Xie, L.; Yang, L.; Hu, Z.; Wang, L.; Wang, Y.; Li, Q.; Guo, S.; Tao, S.-c.; Jiang, H.
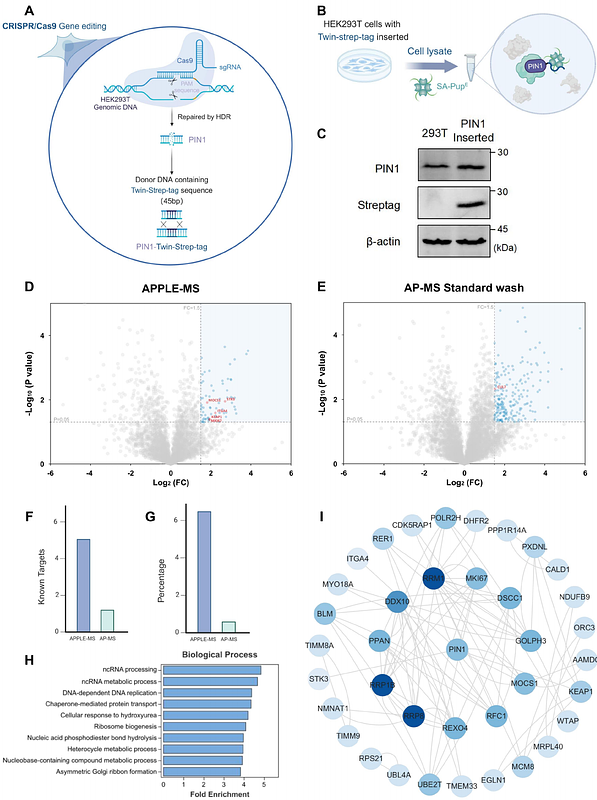
AbstractWhile affinity purification-mass spectrometry (AP-MS) has significantly advanced protein-protein interaction (PPI) studies, its limitations in detecting weak, transient, and membrane-associated interactions remain. To address these challenges, we introduced an innovative proteomic method termed Affinity Purification coupled Proximity LabEling-Mass Spectrometry (APPLE-MS), which combines the high specificity of Twin-Strep-tag enrichment with PafA-mediated proximity labeling. This method achieves unprecedented sensitivity while maintaining high specificity (4.07-fold over AP-MS). APPLE-MS also revealed the dynamic mitochondrial interactome of SARS-CoV-2 ORF9B during antiviral responses, while endogenous PIN1 profiling uncovered novel roles in DNA replication. Notably, APPLE-MS enabled in situ mapping of GLP-1 receptor complexes, demonstrating its unique capabilities for membrane PPI studies. This versatile method advances interactome research by providing comprehensive, physiologically relevant PPI networks, opening new opportunities for mechanistic discovery and therapeutic targeting.