scExploreR: a flexible platform for democratized analysis of multimodal single-cell data by non-programmers
scExploreR: a flexible platform for democratized analysis of multimodal single-cell data by non-programmers
Showers, W.; Desai, J.; Gipson, S. R.; Engel, K. L.; Smith, C.; Jordan, C. T.; Gillen, A. E.
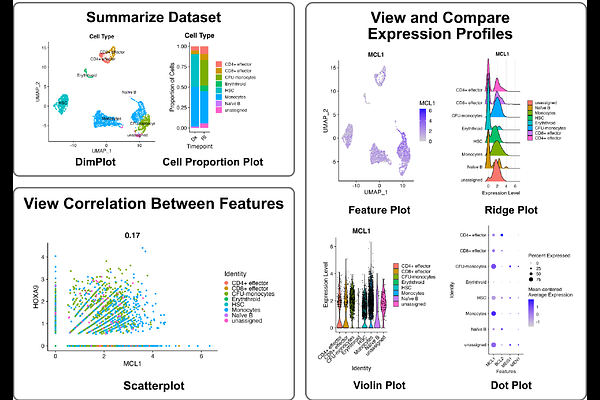
AbstractSingle-cell sequencing has revolutionized biomedical research by uncovering cellular heterogeneity in disease mechanisms, with significant potential for advancing personalized medicine. However, participation in single-cell data analysis is limited by the programming experience required to access data. Several existing browsers allow the interrogation of single-cell data through a point-and-click interface accessible to non-programmers, but many of these browsers are limited in the depth of analysis that can be performed, or the flexibility of input data formats accepted. Thus, programming experience is still required for comprehensive data analysis. We developed scExploreR to address these limitations and extend the range of analysis tasks that can be performed by non-programmers. scExploreR is implemented as a packaged R Shiny app that can be run locally or easily deployed for multiple users on a server. scExploreR offers extensive customization options for plots, allowing users to generate publication quality figures. Leveraging our SCUBA package, scExploreR seamlessly handles multimodal data, providing identical plotting capabilities regardless of input format. By empowering researchers to directly explore and analyze single-cell data, scExploreR bridges communication gaps between biological and computational scientists, streamlining insight generation.