NetREm: Network Regression Embeddings reveal cell-type transcription factor coordination for gene regulation
NetREm: Network Regression Embeddings reveal cell-type transcription factor coordination for gene regulation
Khullar, S.; Huang, X.; Ramesh, R.; Svaren, J.; Wang, D.
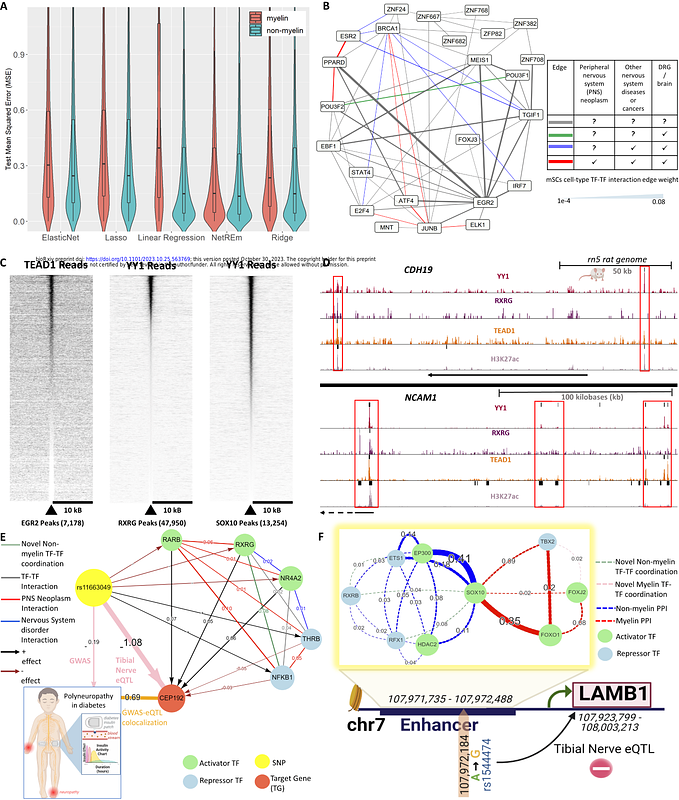
AbstractTranscription factor (TF) coordination plays a key role in gene regulation such as protein-protein interactions (PPIs) and DNA co-bindings. Single-cell technologies facilitate gene expression measurement for individual cells and cell type identification, yet the connection between TF coordination and gene regulation of various cell types remains unclear. To address this, we have developed a novel computational approach, Network Regression Embedding (NetREm), to reveal cell-type TF-TF coordination activities for gene regulation. NetREm leverages network-constrained regularization using prior interaction knowledge (e.g., protein, chromatin, TF binding) to analyze single-cell gene expression data. We test NetREm by simulation data and apply it to analyze various cell types in both central and peripheral nerve systems (PNS) such as neuronal, glial, and Schwann cells as well as in Alzheimers disease (AD). Our findings uncover cell-type coordinating TFs and identify new TF-target gene candidate links. We also validate our top predictions using Cut&Run and knockout loss-of-function expression data in rat and mouse models and compare with additional functional genomic data including expression quantitative trait loci (eQTL) and Genome-Wide Association Studies (GWAS) to link genetic variants to TF coordination. NetREm is open-source available at https://github.com/SaniyaKhullar/NetREm.